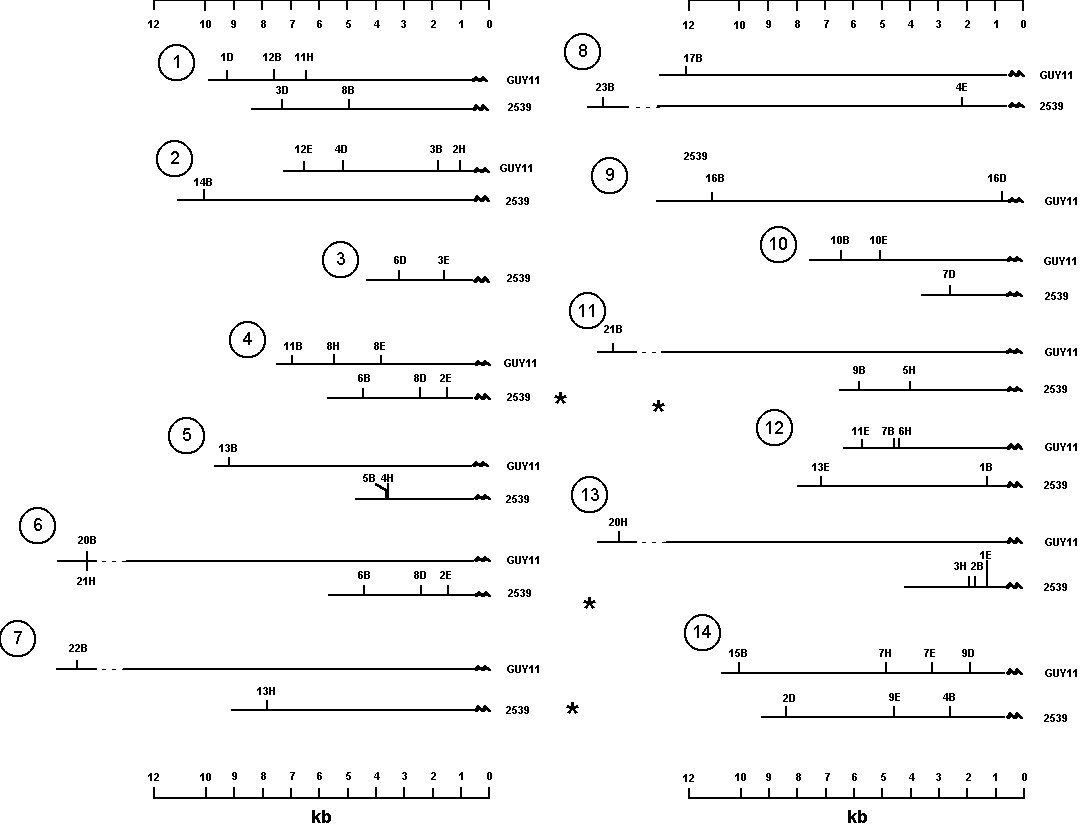
Figure 1: Restriction maps of telomeres of Magnaporthe grisea strains Guy11 and 2539.
Mark L. Farman(1) and Sally A. Leong(1,2) - (1) Department of Plant Pathology and (2) USDA-ARS Plant Disease Resistance Research Unit, Russell Laboratories, 1630 Linden Drive, Madison, WI 53706-1598.
At least five genetic maps exist for the rice blast fungus, Magnaporthe grisea (Romao and Hamer 1991 Proc. Natl. Acad. Sci. USA 89:5316-5320; Sweigard et al. 1993 in Genetic Maps, O'Brien ed. Cold Spring Harbor Press pp. 3.112-3.115; Skinner et al. 1993 Theor. Appl. Genet. 87:545-557; Hayashi and Naito 1993 in Abstracts for International Symposium on Rice Blast Disease, Madison, WI; and Tharreau et al. 1994 in Abstracts of 7th meeting of International Program on Rice Biotechnology, Bali, Indonesia). We have been integrating three of these maps by placing markers used in other laboratories on the map constructed in our laboratory (Skinner et al. 1993, ibid.). The integrated map will be a primary reference map for this fungus and the Guy11 and 2539 parents used to generate the mapping progeny will become standard strains. Guy11 is a natural isolate; therefore this map provides information on the genome organization of a wild, rice-infecting isolate.
As part of a continuing effort to improve the map, the telomeres were mapped to delimit the ends of the chromosomes genetically (Farman and Leong, Genetics, in press). Telomeric restriction fragments of Magnaporthe grisea strains Guy11 and 2539 were identified by hybridizing Southern blots of restricted genomic DNAs with a synthetic oligonucleotide probe (5' TTAGGGTTAGGGTTAGGG 3'). Several restriction fragments hybridized to the oligonucleotide probe and most of these fragments were polymorphic between the two strains. Segregation of telomeric RFLPs generated by four restriction enzymes, BamHI, DraI, EcoRI and HindIII, was examined in 61 progeny of a cross betweeen Guy11 and 2539. As expected, the RFLPs mapped to the ends of linkage groups in the existing genetic map of M. grisea (Skinner et al. 1993, ibid.).
The data set used for mapping was redundant with each telomere being represented by at least two restriction fragments. Note that a telomere marker could be represented by up to eight restriction fragments (4 enzymes X 2 parents). The sizes of the hybridizing fragments revealed the distance from each corresponding restriction site to the end of the chromosome. Therefore, by combining the molecular and genetic analyses, we were able to construct restriction maps for most of the telomeres segregating in this cross (Figure 1). It is important to note that the telomere probe only identifies the most telomere-proximal restriction fragment for each enzyme. As indicated in Figure 1, there are considerable differences in the restriction maps of corresponding telomeres from the two parental strains. In most cases it is not possible to infer simple mechanisms (such as insertions and deletions) to explain this polymorphism since the relative positions of restriction sites with respect to the telomere and to one another are different. Restriction maps of telomeres 4 and 6 from 2539 are identical, indicating that at least one telomere is duplicated in this strain. This conclusion was supported by the observations that the hybridizing band was twice as intense as most other telomeric fragments and that the segregation of the band (presence:absence) was 3 :1. We were not able to produce restriction maps for telomeres 3 of strain Guy11 and 9 of 2539 since we did not identify clearly segregating restriction fragments representing these chromosome ends.
These restriction maps of the M. grisea telomeres provide a useful reference for cloning telomeres or telomere-associated genes from these strains. For example, we used this map to clone Guy11 telomere 2 by ligation-anchored PCR (Farman and Leong, Genetics, in press). The maps are useful for confirming strain identity and for monitoring inheritance of telomeres in crosses involving these parental isolates or their progeny. The genetic map developed by Sweigard et al. 1993 (ibid.) is based on a cross using progeny #6043 from a Guy11 X 2539 cross (Leung et al. 1988 Phytopathology 78:1227-1233).

Figure 1: Restriction maps of telomeres of Magnaporthe
grisea strains Guy11 and 2539.
Positions of sites were determined by measuring the size of the hybridizing fragments generated by each restriction enzyme. Restriction sites are indicated by a number followed by an abbreviation for the restriction enzyme. B=BamHI; D=DraI; E=EcoRI; and H=HindIII. For the BamHI, EcoRI and HindIII restriction sites, the numbers indicate the relative size of each fragment with 1 being the smallest. For example, 3B is the 3rd smallest BamHI fragment. The DraI restriction fragments were numbered in reverse order with 1 being the largest. The reason for this was that, contrary to the other three digests, most restriction fragments were very small and unresolvable. Dotted lines indicate that a fragment was larger than the 12kb marker and the size was therefore estimated. Numbers in circles indicate which telomere each map (or pair of maps) represents. Telomeres that are duplicated in strain 2539 are indicated by *.