The effect of DNA structure and restriction enzymes on transformation efficiencies in
Neurospora crassa
Kristina Garnand and Mary Anne Nelson - Department of Biology, University of New
Mexico, Albuquerque, NM 87131
Addition of the appropriate restriction enzyme to linearized transforming DNA has
been shown to increase transformation efficiencies in organisms as diverse as Saccharomyces
cerevisiae (Schiestl and Petes 1991 Proc. Nat. Acad. Sci. USA 88:7585-7589) and
Dictyostelium discoideum (Kuspa and Loomis 1992 Proc. Nat. Acad. Sci. USA 89:8803-
8807). This process has been described as REMI, for restriction enzyme-mediated
integration. We have examined the effect of restriction enzyme addition on transformation
efficiencies in Neurospora crassa. The frequency of cotransformation of a qa-2 inl double
mutant with two plasmids [one containing the selectable marker qa-2+ (quinate utilization)
and the other containing inl+ (inositol)] was also examined, as was the generation of stable
versus abortive transformants.
Circular (plasmid) DNAS, linear DNAs and linear DNAs plus the restriction enzyme
that had been used to linearize the plasmids were used in these transformation experiments.
The PMSNI plasmid contained the selectable qa-2+ marker (Nelson and Metzenberg 1992
Fungal Genet. Newsl. 39:59-60). The pOKE01 plasmid contained the selectable inl+
marker in an 8.5 kbp PstI insert in pUC18; this plasmid was a gift from Dr. R.L.
Metzenberg. Plasmid DNAs were prepared using the Qiagen, Inc. Plasmid Midi kit. The
restriction enzymes in the digested DNA samples were inactivated by heating at 60 C for
10 minutes.
Preparation of spheroplasts [from a qa-2 aro-9 inl al-2 a (RLM 63-01, a gift of Dr.
R.L. Metzenberg) strain of N. crassa] and their transformation were as previously described
(Schweizer et al. 1981 Proc. Nat. Acad. Sci. USA 78:5086-5090). For the single
transformation experiments, 1 ug of DNA (circular or linear) was mixed with 100 ul
spheroplasts; where noted, 1 ul of the appropriate restriction enzyme, at 15 U/ul, was added
to the linear DNAs immediately prior to transformation. In cotransformation experiments,
a total of 2 ug DNA (1 ug each plasmid), plus 15 U each enzyme (where specified) was
added to 100 ul spheroplasts. Transformants were regenerated in liquid medium and spread
onto selection plates (Akins and Lambowitz 1987 Cell 50:331-345). In the cotransformation
experiments, the regenerated spheroplasts were divided and spread onto plates selective for
qa-2+, inl+ or both qa-2+ and inl+.
Since different preparations of spheroplasts often give quite dissimilar transformation
efficiencies, we controlled for chance variations in transformation efficiencies by doing all
transformations for a given experiment at the same time with a single preparation of
spheroplasts. The results of a typical experiment are shown in Figures 1 and 2. The use of
linear instead of circular transforming DNA did not significantly increase the generation of
stable transformants (Fig. 1 and 2). The addition of restriction enzymes to the transforming
DNA did not have a dramatic or consistent effect in transformations with single DNAs.
Also, transformation with linear and circular DNAs gave approximately the same efficiencies
of cotransformation of the qa-2+ and inl+ markers. However, the addition of restriction
enzymes in the cotransformation experiments resulted in the generation of fewer singly and
doubly-transformed strains (Fig. 1). The reason for the reduction in transformation
efficiencies in cotransformation experiments when restriction enzymes were added is
unclear; the enzymes do not destroy the selectable markers that were used.
The effect of DNA structure on the generation of abortive transformants is shown
in Fig. 2. Abortive transformants are defined as those which begin to grow and form small
colonies on the selection plates, but stop growth prior to making a large colony; these
transformants fail to grow upon transfer, and are thought to reflect cases in which the
transforming DNA enters the cell but fails to become stably integrated into the genome.
The use of linear instead of circular DNA caused a great reduction in the numbers of
abortive transformants that were obtained; the addition of restriction enzyme did not have
a significant effect on the stable/abortive ratio.
In N. crassa the addition of restriction enzymes to linearized transforming DNA was not
shown to increase the efficiency of transformations consistently. This is in contrast to results
obtained with S. cerevisiae and D. discoideum. Although viability was not directly measured,
the similar numbers of transformants obtained when single transforming DNAs were used
suggests that the addition of PstI or HindIII is not resulting in decreased viability. DNA
structure had no significant effect on the frequency of cotransformation with the selectable
markers that were examined. However, the use of linear instead of circular transforming
DNA was shown to significantly decrease the background of abortive (unstable)
transformants.
Supported by NIH Grant GM47374. K.G. acknowledges support from the Howard Hughes
Medical Institute and the Minority Engineering, Mathematics and Science Program at the
University of New Mexico.
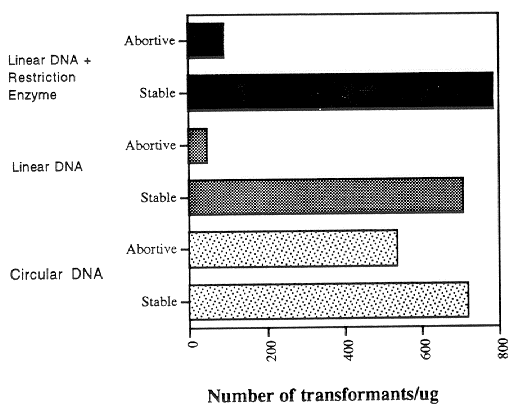
Figure 1. The effect of DNA structure and addition of restriction enzymes on transformation
and cotransformation efficiencies. The numbers of stable transformants obtained per ug
DNA are shown. In the cotransformation experiments, 1 ug each DNA (2 ug total) was
used for the same volume (100 ul) spheroplasts, so the numbers of transformants are per
ug each DNA. In the Transforming DNA column, the symbols are as follows: Co,
cotransformation; L, linearized plasmid DNA; C, circular plasmid DNA; L+RE, linearized
plasmid DNA plus restriction enzyme; inl+, transformation with the selectable inl+ marker
(pOKE01); qa-2+, transformation with the selectable qa-2+ marker (pMSN1). The pMSN1
plasmid was linearized with HindIII, and the POKE01 plasmid with PstI.
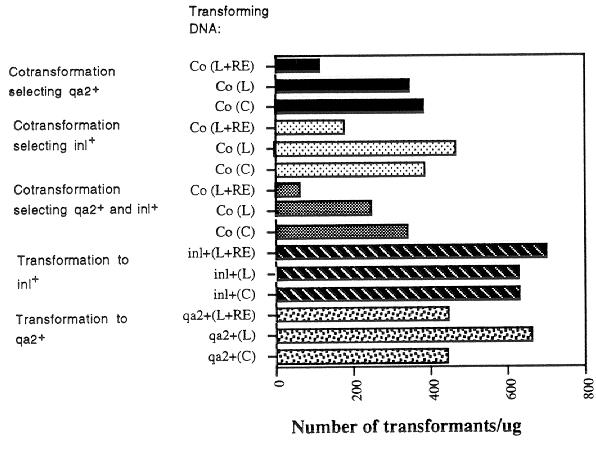
Figure 2. The effect of DNA structure on the generation of stable and unstable (abortive)
transformants. The qa-2 aro-9 inl al-2 a spheroplasts were transformed to inl+ with circular
or PstI linearized forms of the POKE01 plasmid, and the restriction enzyme PstI was added
as noted.

