The Wilson-Garnjobst heterokaryon incompatibility tester strains of Neurospora
crassa contain modifiers which influence growth rate of heterokaryons and
distort segregation ratios.
D.J. Jacobson(1), J. Ohrnberger(2), and R.A. Akins(2) - (1)Department of Botany and Plant Pathology, Michigan State University, East Lansing, MI 48824-1312 (2)Department of Biochemistry, School of
Medicine, Wayne State University, Detroit, MI 48201
Recent interest and accelerated research into the genetics of heterokaryon
incompatibility (HI) in Neurospora crassa has led to increased use of the
original Wilson-Garnjobst HI tester strains available from FGSC (1994 Catalog of
Strains, Part VII.D.1.). We have found inconsistencies and abnormalities in both
growth of heterokaryons and segregation of markers in crosses using these strains.
First noticed was a lack of vigor and incomplete complementation of markers in
forced heterokaryons when compared to compatible heterokaryons with known Oak Ridge
(OR) background. Secondly, skewed allele ratios were recorded in crosses between
the Wilson-Garnjobst strains and strains with OR background. Perkins and Bjorkman
raised a cautionary note about these strains (1978 Neurospora Newsl. 25:24-25), however, they concentrated primarily on the scot mutant present in these
and other strains originating from the Rockefeller-Lindegren (RL) background. We
have attempted to further characterize the erratic behavior of Wilson-Garnjobst
strains and determine if the scot mutant or other modifiers of HI are responsible.
To date, all Wilson-Garnjobst strains tested by us have show the scot
phenotype at 39°C, confirming results of Perkins and Bjorkman (Figure 1). FGSC
1527 is the only strain listed in Part VII.D.1. that was not expected to contain
scot because it is of OR rather than RL background; this proved to be true (data not
shown).
Linear growth rate in race tubes was used to quantify and compare heterokaryon
formation among the strains tested. Loopfuls of conidia from two strains were co-
inoculated at one end of a 25 ml disposable pipet containing 15 ml Vogel s minimal
medium N. (A similar method is described by White and Woodward 1995 Fungal Genetics
Newl. 42) The experiment was limited to the Wilson-Garnjobst strains
that differ at het-c (Table 1); the am1 ad-3B cyh-1R helper strain (FGSC 4564) was included as an OR background
standard.
Strains of like mating type with complementary markers were co-inoculated in all
possible combinations. No attempt was made to control nuclear ratio. Single
strains inoculated on race tubes were used as controls. All tubes were incubated at
34°C and each combination of strains was tested three times. The starting point was
marked as the leading edge of the colony after overnight growth; measurements were
recorded at 24 hr intervals (Figure 2). Although scot is reported to have a
slightly abnormal phenotype at 34°C, it is erratic and difficult to see on
Vogel s minimal medium N (Perkins and Bjorkman 1978 Neurospora Newsl. 25:24-
25). We saw no evidence of the scot phenotype in these experiments.
Heterokaryon formation was confirmed by testing hyphal tips for prototrophic growth.
The leading margin of the colony from the race tube was transferred to a fresh plate
of minimal medium. These cultures were incubated for 1 day at 34°C and 10
single hyphal tips from the colony were subcultured on to separate 10 x 75 mm tubes
of minimal medium. Prototrophic growth in any of the 10 tubes, scored at 2 days,
was considered a positive heterokaryon. The results of hyphal tip subculturing is
indicated on Figure 2 with either a (+) or (-) associated with the line of each
combination tested.
The combination of strains with known OR background, FGSC 1527 and FGSC 4564,
consistently gave the fastest growth; we consider this characteristic of a strong
positive heterokaryon. All other combinations, whether alike or different at
het-c, showed both slower or stalled growth and inconsistency among
replications (Figure 2). Only FGSC 1527 + FGSC 1438, het-C + het-C,
grew the entire length of the race tube (25 cm). However, this happened only twice
in the three replications, with one replication growing less than 1 cm in 9 days.
Moreover, FGSC 1438 + FGSC 4564, the other OR strain, never grew more than 5 cm.
This was generally true of the other het-Cde strains with FGSC 4564 (data not
shown).
Few combinations other than FGSC 1527 + FGSC 4564 yielded heterokaryotic hyphal
tips, including those with like het-c alleles. Correlated with this was
apparent incomplete complementation of the al-2 mutant seen as either white
or mixed white and orange conidia along the length of the race tube. We interpret
these two results as either failure to establish a true heterokaryon or maintain a
stable heterokaryon during continued growth in the race tube. Of interest is the
one het-c + het-C pairing that appeared to form a heterokaryon. This
was probably due to escape from incompatibility.
Additional growth rate tests were performed with other Wilson-Garnjobst strains at
30°C. One set was inoculated with equal number of conidia in an attempt to
control for nuclear ratios. All these additional tests gave similar results as
described above (data not shown).
The growth curves in our Figure 2 resemble the seven types of heterokaryotic growth
reported by Holloway (1955 Genetics 40:117-129, see his Figure 1). In fact,
Holloway used some strains from the Garnjobst collection in his study. In addition,
personal communication from James F. Wilson to David D. Perkins (an unpublished
letter) states, "The inos testers do show a
single gene difference in the ability to form a heterocaryon which will grow the
length of a growth tube, but it is not C, D, or E, does not involve cytoplasmic
incompatibility and does not affect heterocaryon test on slants." Crosses were
performed to determine if het-like genes (such as Holloway s W,
X, Y, and Z) were responsible for the growth curves observed,
especially the reduced vigor of OR and Wilson-Garnjobst het-Cde combinations.
Four Wilson-Garnjobst strains were crossed with multiply marked
centromere (multicent) linkage testers (Table 1), which are OR background and fully
heterokaryon compatible with FGSC 4564. Progeny were scored for all markers and
compatibility with the OR parental background (Table 2). Heterokaryon formation was
tested by pairing all progeny containing a forcing marker with FGSC 4564 in 10 x
75 mm tubes of minimal medium. The difference between strong positive
compatibility and weak or no prototrophic growth was clear at 2 days. This trait
segregated in a 1:1 ratio in the three crosses homozygous for het-C. Alleles
of at least one other marker in each cross showed deviation from the expected 1:1
ratio, affecting linkage calculations from these data. Skewed allele ratios were
also seen when strains derived from the Wilson-Garnjobst strains were crossed with
other strains of OR background (compatible with FGSC 4564) and in one cross between
Wilson-Garnjobst inl and pan-1 strains (data not shown). No marker
was consistently skewed in any of these crosses when repeated. However, het-c and
mt (the only scorable markers) were not skewed in crosses between pan-1 strains
(data not shown).
Of particular interest is linkage between compatibility with OR background and other
markers, as this should indicate approximate location of gene(s) affecting
compatibility. The clearest instance of linkage to OR compatibility is inl
in cross 1027. However, this is complicated by the skewed allele ratio of acr-2.
Both inl and at (linkage group V) show linkage to acr-2 (III)
and as such OR compatibility also appears linked to acr-2. In contrast, cross 1028
shows an apparent linkage between OR compatibility and each ylo-1 (VI) and wc-1
(VII). Again, a skewed allele ratio of wc-1 confounds this analysis. Unlike
cross 1027, there is no apparent linkage between OR compatibility and linkage group
III or V markers.
An unexplainable excess of recombinant progeny is seen between OR compatibility and
each pan-1, psi, and wc-1 in crosses 1030 and 1034. If the
locus modifying compatibility with OR is independent of het-c, as indicated by the
lack of linkage to arg-5, the expectation for the cross heterozygous at
het-c would be a 1:3 ratio of OR compatible to OR incompatible. The data
reveal an opposite ratio of 3:1 compatible:incompatible. This does not correlate
with the observed skewed ratio of mating type. At present, we do not have a good
explanation for the distorted segregation patterns seen in these crosses.
The spreading colonial phenotype of scot is probably not
responsible for the inconsistent growth or incomplete complementation seen, even
though OR compatibility is apparently linked to linkage group V in cross 1027.
First, there is no such linkage in the other three crosses. Second, although
scot is erratic at 34°C, similar results were obtained from a subsample of
pairings repeated at 30°C and 25°C, both permissible temperatures (data not shown).
It is unclear if scot may influence growth rate when heterozygous in
heterokaryons, for example the Wilson-Garnjobst + OR combinations, or if the gene
has a more general pleiotropic effect in modifying heterokaryotic growth. The
linkage
data leave this possibility open, however, other loci on linkage group V or other
linkage groups may also be responsible for modifying compatibility. Since
scot expression may affect growth rate directly, it is also possible that
modifiers of the scot gene cause growth variability in heterokaryons. In any
case, inconsistencies among the pan-1; al-2 strains (crosses 1028, 1030,
1034) and in other repeated crosses does not allow the interpretation of these data
as indicating the location of any loci responsible for skewing or incompatibility
with OR background.
The skewed ratios and apparent linkage of markers on separate chromosomes may
suggest a chromosomal difference between the Wilson-Garnjobst and multicent OR
parents. Although a relatively high number of white inviable ascospores were noted
in the crosses listed in Table 2, there was no regular patterns of black to white
ascospores among shot asci, which would indicate chromosome aberrations (data not
shown). Although we could not completely reject chromosomal differences, presence
of other inviability factors or segregation distorters seems a more likely
explanation.
To further disseminate this information and warn workers of possible complications
using the Wilson-Garnjobst strains, we have requested that the following statement
be added to Part VII.D.1. of the FGSC Catalog: These strains contain scot
and probably other genes in their background that affect both growth of
heterokaryons and segregation of markers in crosses. Heterokaryon tests should be
perfomed at permissible temperature (25°C) and caution should be exercised when
using these strains for genetic studies.
These results also raise the issue of standardizing definitions of compatibility and
incompatibility. This is becoming more important as workers proceed with the
molecular dissection of HI. There is a need to further identify these factors which
may modify the reactions of het genes at the molecular, cellular, and colony
level. In an effort to facilitate standardization in future studies of HI in N.
crassa we suggest that 1) OR background (het-Cde and OR alleles at
het-6 through het-10) be considered the standard against which HI is
measured (FGSC 4564 is especially useful in this context, see Perkins 1984
Neurospora Newsl. 31:41-42); 2) genes of interest (het
genes, modifiers, suppressors, etc.) be crossed into OR background to measure their
effect; and 3) characterizations of compatibility or incompatibility be quantified,
when necessary, by growth rates in race tubes.
Acknowledgments: This work was supported in part by MSU All-University Research
Initiation Grant to DJJ and NIH Award GM43309 to RAA.
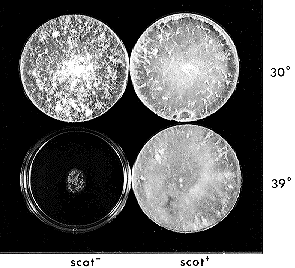
Figure 1. Phenotype of scot at permissive (30°C) and restrictive
(39°C ) temperatures. Plates show 3 days growth on Vogel s minimal medium N
supplemented with pantothenic acid where required. Scot- strain is
FGSC 2659 -- Wilson-Garnjobst het-cDe; pan-1; al-2 a. Scot+ is FGSC
6714 -- Oak Ridge background, T(VL )MB67.

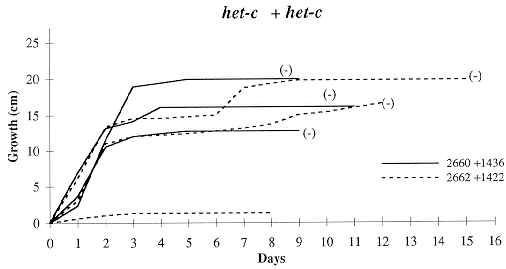
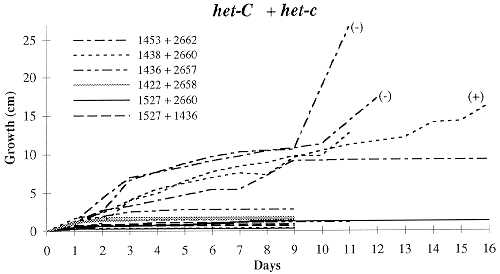
Figure 2. Growth rates of forced heterokaryons in race tubes at 34 C. The het-c alleles of
each pair is indicated at the top of each graph. Each line represents a pair of strains;
each pair was tested three times. Pairings tested for heterokaryotic hyphal tips have a symbol
in parentheses at the end of the line, with (+) indicating prototrophic hyphal tips recovered
and (-) indicating only auxotrophic hyphal tips recovered (see text for details).
Table 1. Strains used for heterokaryon growth rate tests illustrated in Fig. 1 and crosses
listed in Table 2.
het Mating
genotype type Markers FGSC Number
het-Cde A inl 1453
het-Cde A pan-1;al-2 2658
het-Cde a inl 1438
het-Cde a pan-1;al-2 2657
het-Cde a arg-12 1527
het-Cde am1 ad-3B cyh-1R 4564
het-Cde a multicent-4
(arg-5;acr-2;psi; 6829
at;ylo-1;wc-1)
het-Cde A multicent-4
(arg-5;acr-2;psi; 6828
at;ylo-1;wc-1)
het-cde A inl 1422
het-cde A pan-1; al-2 2662
het-cde a inl 1436
het-cde a pan-1; al-2 2660
Note: All strains are Wilson-Garnjobst heterokaryon
incompatibility testers except FGSC 1527, 4564, 6828, and
6829. They are, respectively, an arg-12 mutant, a mating-
type mutant with inactive heterokaryon incompatibility
function of the mating type gene, and the two multiply
marked centromere testers; these strains are all Oak Ridge
background.
Table 2. Allele ratios of markers and apparent linkage among markers in crosses
of Wilson-Garnjobst testers and multiply marked centromere (multicent) testers.
Cross 1027: het-Cde; inl A 1453 X multicent-4 a 6829
Allele Ratios of Markers
mt arg-5 acr-2 psi at ylo-1 wc-1 inl OR(a)
A:a +:- +:- +:- +:- +:- +:- +:- +:-
54:50 48:56 74:30* 48:56 51:53 58:46 60:44 51:53 46:45
Linkage Among Markers
inl & at inl & acr-2 at & acr-2 inl & OR acr-2 & OR
Parentals 65* 75* 65* 80* 62*
Recombinants 38 29 39 11 29
Cross 1028: het-Cde; pan-1; al-2 A 2658 multicent-4 a 6829
Allele Ratios of Markers
mt arg-5 acr-2 psi at ylo-1 wc-1 pan-1 al-2 OR
A:a +:- +:- +:- +:- +:- +:- +:- +:- +:-
26:36 26:36 32:30 38:24 28:34 18:16 27:7* 33:29 34:28 28:30
Linkage Among Markers
mt & al-2 psi & pan-1 ylo-1 & OR wc-1 & OR
Parentals 40* 51* 22* 22*
Recombinants 22 11 10 10
Cross 1030: het-Cde; pan-1; al-2 a 2657 multicent-4 A 6828
Allele Ratios of Markers
mt arg-5 acr-2 psi at ylo-1 wc-1 pan-1 al-2 OR
A:a +:- +:- +:- +:- +:- +:- +:- +:- +:-
66:55 65:56 52:69 63:58 67:54 44:22* 33:31 64:57 64:57 53:68
Linkage Among Markers
mt & al-2 psi & pan-1 pan-1 & OR psi & OR wc-1 & OR
Parentals 77* 101* 46* 37* 23*
Recombinants 44 20 76(b) 84(b) 42(b)
Cross 1034: het-cde; pan-1; al-2 a 2660 multicent-4 A 6828
Allele Ratios of Markers
mt arg-5 acr-2 psi at ylo-1 wc-1 pan-1 al-2 OR
A:a +:- +:- +:- +:- +:- +:- +:- +:- +:-
63:38* 54:47 50:51 49:52 57:44 25:27 28:24 55:46 52:49 71:30*
Linkage Among Markers
mt&al-2 psi&pan-1 pan-1&OR psi&OR mt&OR al-2&OR arg-5&OR
Parentals 60* 82* 39* 36* 61 54 49
Recombinants 41 19 62(b) 55(b) 40 47 52
Notes: Strain numbers are FGSC numbers. Number of progeny tested are listed for
each ratio; number of progeny tested for ylo-1 and wc-1 were limited to albino+
progeny where al-2 was present in one parent. An (*) for any ratio indicates
deviation from a 1:1 ratio at a 5% significance level. Other unlisted linkage
tests betwen marekers are all not significantly different from 1:1. Marker
locations are: mt, linkage group I; arg-5, II; acr-2, III; psi, IV; at, V; ylo-1,
VI; wc-1, VII; inl, VR; pan-1, IVR; al-2, IR.
(a)OR signifies Oak Ridge het background (multicent parent), with (+) indicating
heterokaryon compatibility and (-) indicating incompatibility with FGSC 4564 (am1
ad-3B cyh-1R). Number of progeny tested were limited to those containing an
appropriate forcing marker.
(b) Ratios with unexplained excess of Recombinant progeny.



