Transformations using certain vector/host combinations of Fusarium oxysporum have been shown to promote in vivo production of linear, autonomously replicating plasmids (Powell and Kistler, 1990, J. Bact. 172:3163-3171). The modifications to the transforming vector included: linearization of a circular plasmid, addition of putative fungal telomeres to one end of the plasmid, and duplication and translocation of the modified DNA to the other end of the plasmid. This report confirms that the putative telomere repeats (TTAGGG) from one of these fungal-modified vectors, pFOLT4R4, are also located at the ends of F. oxysporum chromosomes and indicates that these telomere sequences were added to the vector by a recombinational event.
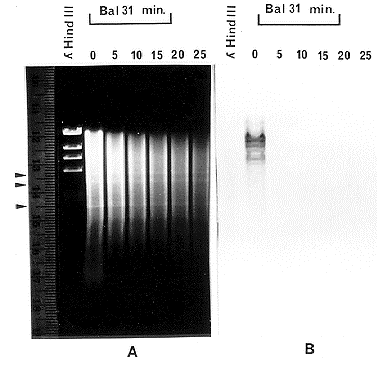
To confirm the putative telomere repeats were located on the ends of F. oxysporum chromosomes, total DNA from F. oxysporum f.sp. conglutinans (strain PHW808) was digested for increasing time intervals with the exonuclease, Bal31. Then, the Bal31 was removed by phenol/chloroform extraction and the DNA was digested with EcoRI and separated on an agarose gel, blotted, and probed with 32P labeled telomere repeats (Figure 1). In panel A of Figure 1, a clear DNA banding pattern was seen which was maintained throughout the digestion. This banding pattern acted as a control demonstrating that internal sequences were protected from Bal31 digestion due to their location away from the chromosomal ends. In panel B, the hybridization signal seen at 0 time was lost within the first 5 minutes of exonuclease digestion indicating the telomere repeat sequences are located at or very near the ends of the chromosomes. Because of these results and due to the sequence similarity of these repeats to other known fungal telomeres (Schechtman, 1989, Fung. Genet. Newl. 36:79-81), it was concluded that these sequences represent the telomere repeats located at the ends of F. oxysporum chromosomes.
To help elucidate the mechanism by which these sequences were acquired by a transforming vector, DNA sequences adjacent to the telomere repeats from pFOLT4R4 and two strains of F. oxysporum were compared to each other and to pUC12 sequences at the site of the in vivo modification. The telomere ends of F. oxysporum f.sp. conglutinans (strain PHW808) and F. oxysporum f.sp. raphani (strain PHW699) were obtained by digestion of their DNA with Sau3A1, amplification of fragments containing the telomeres using anchored PCR (Roux and Dhanarajan, 1990, Biotechniques 8:48-57) primed with the anchor primer and a telomere primer (CCCTAACCCTAACCCTAA), and cloned into pCR1000 (Invitrogen Corp.). This technique produced many clones, but only a few contained the telomere repeats. The clones that did contain the telomere repeats also contained other Sau3A1 DNA fragments. Therefore to ensure no sequences were used which were not located next to the telomeres, only sequences between the telomere repeats and the first Sau3A1 site were compared. Telomere clones from F. oxysporum f.sp. lycopersici (strain 73) in which pFOLT4R4 had been formed, have not yet been obtained. Sequencing of the clones utilized the Sequenase DNA sequencing kit (USB). The results are shown in Figure 2.
The DNA sequences of the telomere repeats and an adjacent twenty-seven base pairs from the two strains of F. oxysporum were the same except for one base substitution (Figure 2). Interestingly, the two F. oxysporum sequences had a significant similarity to a segment in pUC12 which was the site of in vivo modifications in the formation of pFOLT4R4. This DNA sequence similarity could have acted as a recognition site for the host's telomerase or could have provided enough similarity for a recombination event to have occurred between the transforming vector and the host's chromosomes. In addition to the telomere repeats, pFOLT4R4 also contained a three base pair sequence (box in figure 2) which was not part of the pUC12 sequence, but appeared to be of fungal origin because of its similarity to the sequences in 699 and 808. The small difference was probably due to pFOLT4R4 acquiring its telomere sequences from strain 73. Once a good telomere clone is obtained from strain 73, this can be confirmed. With the information available, it can still be concluded a recombination event must have occurred which added the telomere sequences to the vector. A telomerase could have only added the repeating telomere unit (TTAGGG) but recombination could add the repeats and also some flanking sequences as observed in this case. This would explain the initial addition of telomeres to one end of the transforming vector. Locating, subcloning, and sequencing the modification site at the other end of pFOLT4R4 should give answers about how the first modified end was duplicated and translocated to the other side of the vector. It is expected another recombinational event was the most likely cause.
This phenomenon of in vivo modifications is significant to anyone utilizing fungal transformations. Instability of transformants is a very common occurrence in fungal transformations and there have been many explanations for these observations. The possibility of fungal vectors acquiring the host's ARS and/or telomere sequences by recombination could also explain many of these observations and should be considered when evaluating transformations. If this type of modification is common among different fungi, it would be a valuable source of sequences for constructing autonomously replicating vectors.

Figure 1: Bal31 digested F. oxysporum DNA probed with putative telomere sequences.
Panel A. A 0.7% agarose gel containing DNA digested with Bal31 for 0, 5, 10, 15, 20, and
25 minutes. Arrow heads point to DNA bands representing interior DNA fragments.
Panel
B. Blot of gel in panel A, probed with 32P labeled DNA fragment containing 20 telomere
repeat units.
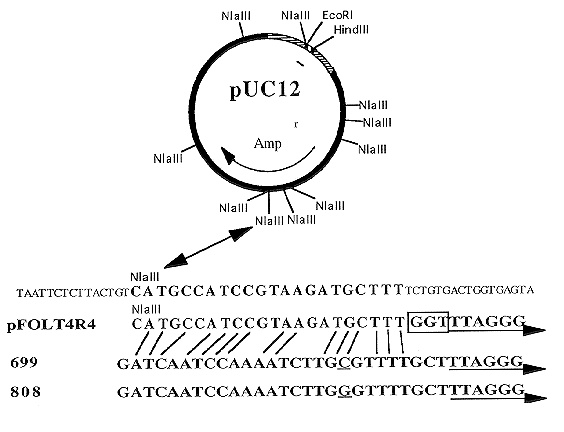
Figure 2: Comparison of sequences from: the in vivo modification site of pUC12; the DNA
adjacent to the telomere repeats of the fungal-modified, autonomously replicating plasmid,
pFOLT4R4; the DNA adjacent to the telomere repeats of F. oxysporum strain PHW699
(699); and the DNA adjacent to the telomere repeats of F. oxysporum strain PHW808 (808).
Arrows underline the beginning of the telomere repeat units which are repeated from three
to eighteen times in the different clones. The box highlights three base pairs in pFOLT4R4
which are not part of the telomere repeats and are not part of the pUC12 sequences. The
underlined bases in 699 and 808 represent the single base difference between these strain's
sequences. The lines between the plasmids (top two sequences) and the telomeres of F.
oxysporum (third and fourth sequence) show one possible sequence similarity.