In vivo site-directed mutagenesis of Neurospora crassa beta-tubulin gene by spheroplasts transformation with oligonucleotides
Mattia Calissano and Giuseppe Macino - Dipartimento di Biopatologia Umana Sezione di Biologia Cellulare, Policlinico Umberto I, Viale Regina Elena 324, 00161 Rome, Italy
The use of Polymerase Chain Reaction (PCR) to incorporate mutated oligonucleotides into newly synthesized segments of DNA and the subsequent introduction of this DNA into competent cells is one of the most widely used methods of site-directed mutagenesis (Zoller et al. 1984 DNA 3:479-488; Dalbodie-McFarland et al. 1982 Proc. Natl. Acad. Sci. USA 79:6409-6413). Several variations of this technique exist, but all require several and some time-consuming steps. Here we report the results of a rapid method of in vivo site-directed mutagenesis involving the direct transformation of Neurospora crassa spheroplasts with mutated oligonucleotides.
In 1986 a mutant of the beta-tubulin gene of Neurospora crassa , resistant to the toxic compound benlate (Benomyl) was isolated (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461). This mutant has a single point mutation that determines a phenylalanine-tyrosine substitution at position 167 of the amino acid sequence. We decided to take advantage of this known mutation to evaluate the feasibility of carrying out in vivo site-directed mutagenesis in Neurospora crassa using mutated oligonucleotides. Our experimental approach involved direct transformation of Neurospora crassa spheroplasts with both sense and antisense oligonucleotides of various lengths, complementary to the mutated beta-tubulin gene.
We synthesized 30, 40 and 50-mer sense and antisense oligonucleotides carrying not only the phenylalanine-tyrosine substitution at position 167, but also a silent point mutation as a marker to determine if the Benomyl-resistant mutants obtained resulted from incorporation of the mutation carried by the synthetic oligonucleotide and not to spontaneous mutations.
108 Neurospora crassa wild type (74-OR23A - FGSC # 987) spheroplasts, prepared according to Schweizer et al. 1981 Proc. Natl. Acad. Sci. USA 78:5086-5090, were transformed in parallel with 5ug of sense and 5ug of antisense oligonucleotides carrying the two mutations, according to a standard Neurospora crassa transformation protocol (Vollmer and Yanofsky 1986 Proc. Natl. Acad. Sci. USA 83:4869-4873); 500 ul aliquots of this cell suspension were plated on sterile Neurospora crassa minimal medium (Davis & De Serres 1970 Methods Enzymol. 17:79-143) supplemented with Benomyl (1ug/ml) and incubated at 29 C for two days.
Only cells transformed with the antisense oligonucleotide gave rise to Benomyl-resistant colonies. These transformants were subjected to at least two passages of purification in slants supplemented with Benomyl (1ug/ml). One-hundred nanograms of the chromosomal DNA of two of these transformants was used as a template in a standard PCR reaction to amplify the beta-tubulin gene region presumably carrying the two mutations. Sequencing of the PCR fragments confirmed that both the phenylalanine-tyrosine substitution and the silent point mutation were present in the transformants.
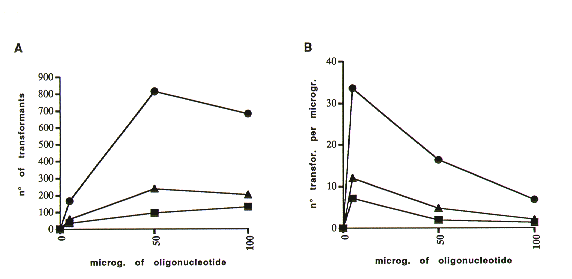
To evaluate whether the efficiency of this mutagen technique depends on the quantity of oligonucleotides used, 108 Neurospora crassa wild type spheroplasts were transformed directly with 5, 50 and 100 umg of the mutant antisense oligonucleotide and plated as described above. Our data (Figure 1) show that while mutagenetic efficiency was directly proportional to the amount of the oligonucleotides used to at least 50 ug, there was an inverse correlation between the number of transformants and length of the oligonucleotide.
This method of in vivo site-directed mutagenesis has also been successfully used on the cyc-1 gene of Saccharomyces cerevisisae (Moerschell et al. 1988 Proc. Natl. Acad. Sci. USA 85: 524-528). In this case, mutagen efficiency was not only directly correlated to the length and amount of the oligonucleotides used (Yamamoto et al. 1992 Yeast 8: 935-948), but was nearly 20-fold more efficient using the sense rather than the antisense oligonucleotide (Yamamoto et al. Genetics 131: 811-819). An intriguing hypothesis to explain the difference in the transformation efficiency between the sense and the antisense oligonucleotide both in Neurospora crassa and Saccharomyces cerevisiae is based on DNA replication; i.e., after oligonucleotides enter into cells they could specifically anneal to the complementary lagging strand as Okazaki fragments during DNA replication. The incorporation of a sense or antisense oligonucleotide would therefore simply reflect the different direction of the replication fork of a gene.
The results reported here clearly show that is possible to perform in vivo site-specific mutagenesis in Neurospora crassa using oligonucleotides. The simplicity and speed of the method described here make it a particularly suitable technique for characterizing any selectable Neurospora crassa gene.

Figure 1. Correlation of the number of Benomyl-resistant colonies and the amount and length of the antisense oligonucleotides used for transformation.
The x asis, in both diagrams, reports the amount of oligonucleotide, expressed in micrograms. The y axis in A reports the number of Benomyl-resistant colonies, and in B the ratio between the number of Benomyl-resistant colonies and the amount of oligonucleotide used. B.30 B.40 B.50 indicate the 30, 40 and 50-mer anti-sense oligonucleotides.
Return to the FGN43 Table of Contents
Last modified 7/25/96 KMC