Dipnath Baidyaroy1 and Helmut Bertrand2 - 1Department of Botany and Plant Pathology and 2Department of Microbiology, Michigan State University, East Lansing, Michigan 48824-1101
Methods for the isolation of highly purified mitochondria (Lambowitz 1979 Meth. Enzymol. 59:421-433) and mitochondrial DNAs (Vierula and Bertrand 1992 Mol. Gen. Genet. 234:361-368) from the filamentous fungi have been available for some time. However, so far, there is no procedure that yields pure plasmid DNAs from mitochondria. We have developed an easy approach for the extraction of circular, mitochondrial plasmids from Neurospora which yields small circular DNAs suited for the analysis of replication intermediates by two-dimensional gel electrophoresis. This method should work well for the extraction of circular mitochondrial plasmids from filamentous fungi in general.
Mycelium was grown in liquid medium for 16-18 h and harvested on Schleicher & Schuell #470 filters in Buchner funnels, washed repeatedly on the filter with distilled water and then once with isolation buffer (0.44 M sucrose, 10 mM Tris-HCl, 5 mM EDTA pH 7.5). Hyphae were disrupted in cold isolation buffer with a grind-mill (Weiss et al. 1970 Eur. J. Biochem. 14:75-82), but grinding with sand can be substituted. The homogenized mycelium was centrifuged for 10 min at 4,000 rpm (1,912 x g) in the cold in a Sorvall SS34 rotor. The supernatant liquid was collected and centrifuged again in the cold for 10 min at 4,000 rpm. The supernatant liquid was again collected in fresh tubes and centrifuged at 4 for 40 min at 16,000 rpm (30,590 x g). The supernatant liquid was discarded and a standard alkaline lysis bacterial plasmid preparation (Sambrook et al. 1989 Molecular Cloning : A Laboratory Manual, Vol.1, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. p.1.25-1.26) was performed on the pellet of crude mitochondria. The pellet was resuspended in 400 ul of a resuspension buffer (50mM Tris-HCl, 10 mM EDTA, 100 ug/ml RNase A, pH 7.5) by gentle vortexing and then transferred into a 1.5 ml Eppendorf tube. Then, 400 ul of cell lysis solution (0.2 M NaOH, 1 % SDS) was added and mixed thoroughly with the suspension of mitochondria. After 5 min of incubation at room temperature, 400 ul of cold neutralization solution (1.32 M potassium acetate, pH 4.8) were added and mixed with the lysed mitochondria by gently inverting the tube. The mixture was kept on ice for 5 min before it was centrifuged at full speed in an Eppendorf microcentrifuge for 25 min in the cold. The supernatant liquid was collected and passed through a column of 66.9 % Guanidine-HCl resin (as supplied by Promega with the WizardTM Minipreps DNA Purification System, Cat. # A7100) and the DNA was processed according to the manufacturer's directions.
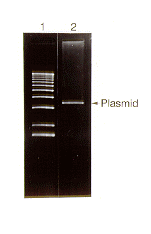
Using this technique, we were able to purify even high molecular weight multimeric forms of mitochondrial plasmids. The yields were reasonably high and the DNA obtained was found to be of good quality (Figure 1) and could be used for standard molecular analyses, including restriction endonuclease mapping and PCR amplification.

Figure 1. Ethidium bromide stained agarose gel showing the electrophoretic migration pattern of a BglII-digested mitochondrial plasmid preparation (lane 2) from the Mauriceville strain of Neurospora crassa. Lane 1 provides molecular weight standards (1-kb ladder).
Supported by grant 95-37303-1785 from the U.S. Department of Agriculture.
Return to the FGN 44 Table of Contents