James D. Colandene and Reginald H. Garrett- Department of Biology, University of Virginia, Charlottesville, Virginia, 22903
We report the isolation of a cDNA clone containing a segment of DNA representing a gene which appears to require the nit-4 gene product for expression, yet is not regulated by nitrate induction. Such a cDNA is a novel finding with respect to the known function of the nit-4 gene.
Expression of the Neurospora crassa nit-3 and nit-6 genes encoding nitrate reductase and nitrite reductase (enzymes of the nitrate assimilatory pathway), respectively, requires simultaneous transcriptional activation by the nit-2 and nit-4 gene products (Exley et al. 1993 J. Bacteriol. 175:2379-2392; Fu and Marzluf 1987 Proc. Natl. Acad. Sci. 84:8243-8247). The nit-2 gene product is a global regulatory element required for the expression of many pathways leading to the acquisition of secondary nitrogen sources when reduced nitrogen sources are scarce (Coddington 1976 Mol. Gen. Genet. 145:195-206). The nit-4 gene product is a pathway-specific transcription factor which exerts positive control of nit-3 and nit-6 in the presence of nitrate (Fu et al. 1989 J. Bacteriol. 171:4067-4070). The only known function of the nit-4 gene product is to activate expression of nit-3 and nit-6 during nitrate induction.
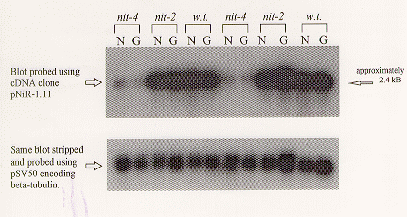
A cDNA clone was identified during an attempt to isolate a full-length nit-6 cDNA from a Lambda ZAPII cDNA library using the radiolabelled nick-translated product of a nit-6 partial cDNA as probe. The clone identified was isolated as a pBluescript phagemid and designated pNiR-1.11. pNiR-1.11 was nick-translated to produce 32P-radiolabelled DNA to probe a nitrocellulose filter of electrophoretically separated polyA+-RNA isolated from wild-type (FGSC#987), as well as nit-2 (FGSC#983) and nit-4 (FGSC#985) mutant N. crassa mycelia grown under glutamine-repressing or nitrate-inducing conditions. Interestingly, the pattern of expression revealed upon exposure of the filter to X-ray film was unlike nit-6 (Figure 1). First of all, the signal was very strong after an overnight exposure. Typically, the signal for nit-6 is weak even after exposing the X-ray film for 24 hours or longer, due to the low abundance of nit-6 message. More significant is the fact that the RNA recognized by this probe did not appear to be regulated in response to nitrate induction in wild-type or nit-2 mutant mycelia. That is, this RNA appeared to be expressed in equal amounts in both wild-type and nit-2 mutant mycelia whether the mycelia were nitrate-induced or glutamine-repressed, unlike nit-6 which is only expressed under nitrate-inducing conditions. Most notable, however is that this RNA was present at far lower levels (almost undetectable) in the nit-4 mutant mycelia under either nitrate-inducing or glutamine-repressing conditions. The blot was stripped and reprobed using nick-translated 32P-radiolabelled insert from cosmid pSV50. This cosmid carries a N. crassa B-tubulin gene, which is constitutively expressed. This probe provides a control to determine the relative amounts of RNA in each lane. The results indicated that nearly equal amounts of RNA had been loaded in each lane (Figure 1).
The pNiR-1.11 cDNA clone apparently represents a portion of a gene that is not regulated by nitrate induction yet requires a functional nit-4 gene for expression. The identification of a nit-4-dependent RNA that is unresponsive to nitrate induction (or glutamine repression) is an unexpected result, since nit-4 is believed to be involved only with the mediation of nitrate induction.
The 5' and 3' ends of this clone were sequenced using the T3 and T7 primers hybridizing to the flanking regions of the polycloning site in the pBluescript vector. The sequence determined at one end of the insert (using the T3 primer) was found to be identical to a sequence within the nit-6 cDNA (pNiR-3), thus explaining why the clone was initially identified using nick-translated pNiR-3 as a probe. The other end, however, showed no homology to nit-6, which explains why a different expression pattern was seen. It seems likely that the ends of two separate cDNAs were ligated together during library construction. The unidentified gene segment within pNiR-1.11 had the following 184-nucleotide sequence.
1 TACTCGACTT TTCCAACCAT CTCTTCCTTA CTCTGTTTAC AATAATACAT 51 CATACCCGAA CCTTCCATAT TCACTACATC TTGTAATAGT CGGTCGAATT 101 CTTTCCACTC TTCCATCTCT GCGACATCAC CTCACACAGC ACATTCATTC 151 AACTTGACGA TTACCACTGA ATAATACCTT CAAC
The program FASTA was used to search Genbank for similarity between this nucleotide sequence and the potential amino acid sequences resulting from translation in either orientation. No similarity close enough to warrant a possible identification of this gene was detected.
If the unidentified gene is regulated by nit-4, it is not solely responsive to nitrate induction, since it appears to be expressed to the same extent under nitrate-inducing or glutamine-repressing conditions in wild-type and nit-2 mycelia. New energy requirements and metabolic changes must occur when an oxidized form of nitrogen such as nitrate is the sole nitrogen source. Therefore, it is plausible that a regulatory element mediating nitrate induction, such as the nit-4 gene product, might also be involved with the regulation of other pathways.
This clone (pNiR-1.11) is being deposited with the Fungal Genetics Stock Center for anyone wishing to pursue the matter further.

Figure 1 Northern analysis of clone pNiR-1.11 expression in wild-type, nit-2 (strain nr37, FGSC #983) and nit-4 (strain nr15A, FGSC #985) mutant N. crassa mycelia. Strains were grown under either nitrate-inducing (N) conditions (20 mM sodium nitrate as sole nitrogen source) or glutamine-repressing (G) conditions (20 mM glutamine as sole nitrogen source). Poly A+-RNA was isolated from mycelia extracts and 10 g (based on A260) loaded in each lane of a formaldehyde agarose gel; duplicate sets of samples were run, as indicated by the repeated pattern. The RNAs were then transferred to nitrocellulose membrane and probed with nick-translated pNiR-1.11. The same blot was stripped and reprobed with the cosmid pSV50, which carries a Neurospora gene for B-tubulin.
Return to the FGN 44 Table of Contents