
Lisbeth Hamer and Shelly Gilger- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907 USA
We have constructed 2 tn5-containing plasmids, pLH1 and pLH3, specialized towards mutagenesis of genes from fungi which are auxotrophic for arginine or sensitive to hygromycin (such as the filamentous fungi Aspergillus nidulans and Magnaporthe grisea.) These plasmids are also a useful means of integrating additional marker genes in the plasmid backbone.
Application of gene knock-out technology in filamentous fungi by homologous recombination generally requires construction of a disruption vector. This vector is composed of parts of the gene flanking an appropriate marker gene. To speed up the process of disruption vector construction, we have combined the transposing ability of tn5seq1 (Nag et al. 1988 Gene 64:135-1145) and the thermosensitive rep gene from pFDX600 (Vogele et al. 1991 Nucl. Acids Res. 19:4377-4385) as described (Urban et al. 1996 Mol. Gen. Genet. 250:414-420) with the argB (Upshall et al. 1986 Mol. Gen. Genet 204:349-354) gene marker for ornithine carbamoyltransferase (pLH1) and the hph (modified, from Carroll et al. 1994 Fungal Genet. Newsl. 41:22-88) gene marker for hygromycin B phosphotransferase (pLH3). The argB and hph marker genes are functional in A. nidulans and M. grisea, respectively. Transposition events can be induced in E. coli cells harboring both the plasmid containing the gene to be disrupted and pLH1/3. Application of high selection pressure against the presence of pLH1/3 (restrictive temperature) and towards the transposition of tn5seq1 (high antibiotic concentration) allows identification of transposition into the target plasmid. Figure 1 outlines the procedure and the mutagenesis protocol is provided at the end of this article.
We show that the 6.1kb DNA fragment containing the sepA gene from A. nidulans strain A28 (pabaA6 biA1) cloned in E. coli (pLH5) (Harris et al. 1997 EMBO submitted) can be readily mutagenized by pLH1. We have made multiple disruption derivatives of pLH5 with the concomitant introduction of the argB gene (Figure 2). Using a 3.8 kb pBR322-derived backbone, transpositions into the 6.1 kb sepA gene were observed at a frequency of 0.25. The exact position of the disruption can easily be determined, due to the presence of SP6 and T7 priming sites at the ends of the intervening tn5seq1/argB cassette. Because the disruption is generated by an insertion, the entire sepA gene is preserved and provides the targeting sequence for later in vivo disruption of the sepA gene. Circular integration events of the sepA/tn5seq1 disruption plasmid in the A. nidulans strain ATW17 (argB) have been verified (Figure 3). A parallel example to the integration of the argB gene (as shown by pLH106) was verified by use of the tn5seq1/hph cassette from pLH3. In order to acquire an additional marker for selection, the hph marker has been transposed into the vector backbone of a plasmid later used for transformation of M. grisea. The function and presence of the hph gene in the M. grisea spores was confirmed (F. Tenjo, unpublished). pLH1 and pLH3 are available at the Fungal Genetics Stock Center.

Bg=BglII, Bc=BclI, P=PstI, Nc=NcoI, A=AvaII, Sm=SmaI, S=SalI, H=HindIII, N=NheI, No=NotI, X=XhoI, Ss=SspI. The BamHI sites flank the transposing cassette. kan= kanamycin resistance gene, tnp =transposase, arg=ornithine carbamoyltransferase gene, hph=hygromycinB phosphotransferase gene
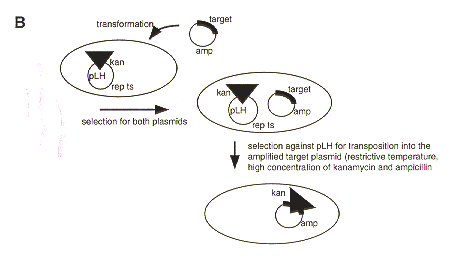
Figure 1. A. Construction of pLH1 and pLH3. pLH1: The 1.55 kb argB gene was amplified with artificial XhoI overhangs and ligated to SalI digested pFTS. pLH3: A 1.4 kb SalI fragment containing the hph gene was ligated to SalI digested pFTS. B. Transposon mutagenesis using pLH1 and pLH3. Competent E. coli cells containing pLH1 or pLH3 are transformed with the plasmid containing a gene to be disrupted. Transformants containing the pLH plasmid and the target plasmid are selected on medium containing ampicillin (selection for target plasmid) and kanamycin (selection for tn5seq1). The temperature and antibiotic concentration are raised to counterselect the pLH plasmid and promote transposition as well as the amplification of the target plasmid. Plasmids that are both kanamycin and ampicillin resistant are characterized and the exact site for tn5seq1 integration determined by restriction analysis (and/or sequencing).
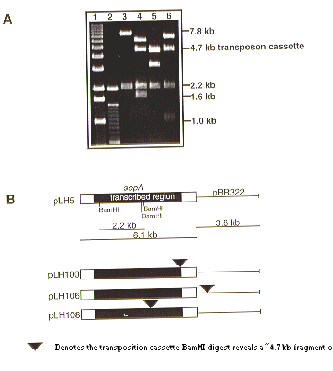
Figure 2. A. Examples of BamHI restriction digests of pLH5 deletion derivatives. Lanes 1: 1 kb lambda ladder (GibcoBRL); 2: 100 bp ladder (GibcoBRL); 3: pLH5; 4: pLH100; 5: pLH106; 6: pLH108. B. Schematic presentation of pLH5 deletion derivatives. pLH5: intact sepA gene, no tn5seq1 transposition; pLH100: transposition in 3’ end of sepA; pLH106: transposition in pBR322 vector backbone; pLH108: transposition in vital part of the sepA gene (see Figure 3).
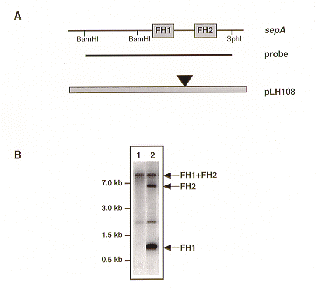
Figure 3. Circular integration of the sepA/tn5seq1 cassette at the A. nidulans sepA locus. A. BamHI/SphI restriction map of the sepA locus, the position of the sepA probe used in panel B, and the disruption vector pLH108. B. Verification of disruption of the sepA locus by Southern analysis. BamHI restriction digest of the parental A. nidulans strain ATW17 (lane 1), and a sepA integration event (lane 2). FH = formin homology.
Protocol: TRANSPOSON MUTAGENESIS USING PLH1 AND PLH3
ASPERGILLUS NIDULANS: USE PLH1 (CONTAINS THE argB gene)
MAGNAPORTHE GRISEA: USE PLH3 (CONTAINS THE hph gene)
1. Make competent E. coli strains LH1 or LH3 at 28 C (provided on agar, LH1 harbors pLH1,
LH3 harbors pLH3).
2. Transform your favorite plasmid (has to carry alternative (to kanamycin) drug resistance gene, i.e., ampicillin resistance) into competent E. coli strain LH1 or LH3. It is important that you are familiar with the BamHI restriction map of your plasmid.
3. Incubate 1 hr in SOC at 28 C.
4. Plate on 1Xkan (25 mg/l), 1Xamp (50mg/l) plates (2XTY) (aliquots of 1 ul, 10 ul, and 100 ul).
5.Pick single colonies to ONC in liquid 4 ml (2XTY) cultures 1Xkan, 1Xamp, 28 C.
6. Plate aliquots of 0.4 ml on 10Xkan, 1Xamp plates (2XTY) incubate for 2 days at 43 C.
7. Pick large colonies, restreak onto 1Xkan, 1Xamp plates (2XTY).
8. ONC in liquid 4 ml (2XTY) cultures 1Xkan, 1Xamp, 37 C.
9. Miniprep colonies
10. Digest with BamHI (gives an approximate 4.7 kb diagnostic band fr the pLH1 transposon and an approximate 4.55 kb diagnostic band for the pLH3 transposon). Map or sequence the insertion.
11. When appropriate disruption is verified, transform back into original host.
12. Select for arginine prototrophy (A. nidulans) or hygromycin resistance (M. grisea).
Common problems:
Sometimes the transposon cassette will integrate into the bacterial genome. This can be determined if the miniprepped plasmids from step 9 can transform kan/amp sensitive E. coli cells to resistance to these antibiotics.
We have noticed a tendency for nearly identical transposition site selection when using pUC and pBluescript plasmids as targets. This is only preferable if no gene desruption but a marker integration into the vector backbone is wanted.
Return to the FGN 44 Table of Contents