Michaël Tognolli, Anne Utz-Pugin, Gilbert Turian and Claude Rossier - Laboratory of Microbiology, University of Geneva, Place de l'Université 3,CH-1211 Genève 4, Switzerland
Mutants of N. crassa highly resistant to benomyl with respect to hyphal growth were obtained by mutagenizing wild type strain St. Lawrence 74A or mutant Bml 511(r) which is moderately resistant to the fungicide. One of these, strain E1-91 that has a mutation mapping at the Bml (beta-tubulin) locus, showed temporary sensitivity to the benzimidazole.
Neurospora crassa is a convenient model to investigate the hypothetical interrelations between the patterns of germ tube emergence and extension and cytoplasmic or spindle microtubules (MTs). This fungus has only one gene for the MT component beta-tubulin (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461), the target for benomyl and one beta-tubulin polypeptide (Hoang-Van et al. 1989 Eur. J. Cell. Biol. 49:42-47). Moreover, mutants resistant to the fungicide have been isolated (Borck and Braymer 1974 J. Gen. Microbiol. 85:51-56). Benomyl resistance shown by these mutants is due to single amino acid substitutions in the beta-tubulin polypeptide (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461; Fujimura et al. 1992 Curr. Genet. 21:399-404). In this article, we describe new mutants that display high resistance to the antitubulin agent benomyl as well as strains that are temporarily sensitive to the benzimidazole.
Methods
Neurospora crassa wild type strain St. Lawrence 74A (FGSC 262), benomyl-resistant strains Bml 511(r) A (FGSC 2965) and strains trp-2; ylo-1; chol-2 A, a (FGSC 4137, 4138) were obtained from the Fungal Genetics Stock Center, University of Kansas Medical Center, Kansas City, KS, USA. Strains were grown at 33 deg.C in Vogel's minimal medium containing 1.5% (w/v) sucrose. Linear growth rate was measured in race tubes according to Ryan et al. (1943 Am. J. Bot. 30:784-799).
Benomyl resistant colonies were selected at 33 deg.C after plating UV-irradiated conidia (75% kill; 106 per 8.5 cm diameter Petri dish) of wild type strain St. Lawrence on sorbose medium containing 1 uM benomyl. Another selection strategy was to plate UV-irradiated conidia of benomyl-resistant strain Bml 511(r) on sorbose medium containing 500 uM benomyl (no growth of mutant Bml 511(r)). The resistant allele in a few mutants highly resistant to benomyl was mapped with respect to markers trp-2, ylo-1 and chol-2 in a cross to the triple mutant strains 4137 and 4138 according to Borck and Braymer (1974 J. Gen. Microbiol. 85:51-56). The tolerance levels to the fungicide were scored at final concentrations of 1, 25 and 250 uM. After analysis of the auxotrophic characters, recombination frequencies between all markers were then determined.
Results and Discussion
Benomyl-resistant mutants were isolated from wild type strain St. Lawrence with a frequency of 6 x 10-6. Mutants like 47 and 72 (Table 1), displaying high resistance to the microtubule inhibitor, represented about 5% of all mutants. Four highly resistant mutants (D2, I3, E1 and J28, Table 1), isolated with a frequency of 5 x 10-7 by mutagenezing benomyl-resistant mutant Bml 511(r), very likely bear a second mutation in the beta-tubulin gene. This is in agreement with the finding that various mutations in this gene confer benomyl resistance (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461; Fujimura et al. 1992 Curr. Genet. 21:399-404). Linear growth of Bml 511(r) is inhibited by 90% at 100 uM benomyl. In contrast, all six highly resistant mutants were only slightly affected (about 35% inhibition) at this high inhibitor level (data not shown).
Multiple germ tube formation is induced by 1 uM benomyl treatments in the wild type (Caesar-Ton That et al. 1988 Eur. J. Cell. Biol. 46:68-79). Interestingly, benomyl-resistant strain J28-48 was found to germinate by multipolar outgrowth in the absence of any benomyl treatment (Table 1).
All mutations conferring high benomyl resistance were found to map at the Bml (beta-tubulin) locus on linkage group VI (Table 2). We found that crosses of mutant E1 to strain trp-2; ylo-1; chol-2 gave rise to clones (1 to 5% of the ascospore progeny) that displayed temporary sensitivity to benomyl (Table 1). One of these, E1-91, germinated by multiple germ tube formation in the presence of 10 uM benomyl. This multipolar outgrowth lasted for about 24 h before a normal linear hyphal extension resumed in the presence of the drug. The high benomyl resistance of mutant E1-91 was also assigned to the Bml locus (Table 2). However the temporary sensitivity phenotype was not transmitted through meiosis: cross of strain E1-91 to strain trp-2; ylo-1; chol-2 gave rise, among the benomyl-resistant ascospore progeny, only to clones like E1-91-1 (Table 1) that displayed normal high resistance to the MT inhibitor.
Table 1. Neurospora crassa strains used in the present study.
Strains Origin / Comments Wild-type St. Lawrence Vegetative reisolate of St. Lawrence 74A. Benomyl-sensitive. Bml 511(r) Phenylalanine-to-tyrosine change at position 167 of beta-tubulin (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461). Moderate benomyl resistance. 47 UV mutagenesis of wild type. High benomyl resistance. 72 UV mutagenesis of wild type. High benomyl resistance. D2 UV mutagenesis of Bml 511(r). High benomyl resistance. I3 UV mutagenesis of Bml 511(r). High benomyl resistance. E1 UV mutagenesis of Bml 511(r). High benomyl resistance. E1-91 Progeny of cross E1 x trp-2; ylo-1; chol-2. Temporary benomyl sensitivity. E1-91-1 Progeny of cross E1-91 x trp-2; ylo-1; chol-2. High benomyl resistance. J28 UV mutagenesis of Bml 511(r). High benomyl resistance. J28-48 Progeny of cross J28 x trp-2; ylo-1; chol-2. Spontaneous multipolar outgrowth.
Table 2. Recombination frequencies in regions 1 to 5 (see partial map of linkage group VI below) as determined from crosses of benomyl-resistant strains with triple mutant strains 4137 and 4138. Regions between the markers utilized in mapping of the Bml allele are arbitrarily numbered. Map distances derived by calculation of mean values are in brackets.
Recombination (%) in region: # Ascospores
Resistant
strains 1 2 3 4 5 analyzed
47 22.2 1.1 23.1 31.1 30.0 90
72 24.0 4.0 28.0 33.0 35.0 100
D2 27.0 5.0 30.0 34.0 39.0 100
I3 19.8 2.1 21.9 29.2 29.2 96
E1 16.5 4.1 20.6 39.1 39.2 97
E1-91 18.0 4.0 22.0 24.0 28.0 100
J28 15.0 1.1 14.0 30.1 29.0 93
J28-48 18.6 4.6 22.1 23.2 26.7 86
Mean 20.1 3.3 22.7 30.5 32.0
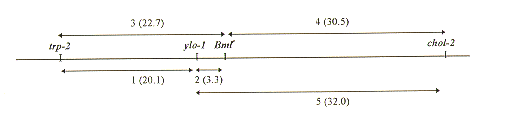
Return to the FGN 44 Table of Contents