A rapid procedure for the extraction of genomic DNA from intact Aspergillus spores
Pieternella C. Mol, Isabelle Mouyna and Christophe d'Enfert* - Laboratoire des Aspergillus, Institut Pasteur, 75724 Paris Cedex 15, France
Genomic DNA of different species of Aspergillus was prepared from intact spores using the Nucleon MiY kit of Amersham. The method is rapid, does not involve mechanical disruption of the spores nor the use of phenol-chloroform extractions and yields DNA that is suitable for PCR amplification and Southern analysis. The method is also applicable to mycelium ground with glass beads.
Preparation of good-quality genomic DNA from fungal species most frequently involves the disruption of the thick cell wall by mechanical grinding or enzymatic lysis followed by the removal of cellular proteins by organic solvents. In this regard, the preparation of a large number of samples that are suitable for further analysis like Southern blotting is somewhat strenuous and time consuming. Procedures that take advantage of chemical lysis and that obviate the need for phenol-chloroform extraction have been devised and successfully applied to the preparation of genomic DNA from different yeast species including Saccharomyces cerevisiae, Schizosaccharomyces pombe and Pichia pastoris. The Nucleon MiY kit available from Amersham (RPN8518) provides a rapid procedure for DNA extraction from yeast minipreps with a total yield of ca. 10 ug/108 cells and DNA that is suitable for Southern analysis. Here, we show that the Nucleon MiY kit can be used successfully to prepare good-quality genomic DNA from conidia from different Aspergillus species.
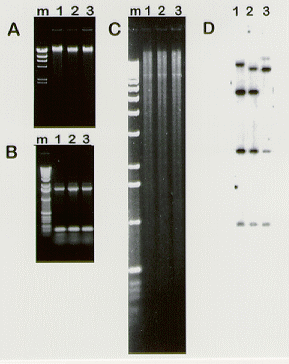
A. fumigatus or A. nidulans strains were sporulated on complete medium (Cove 1966 Biochim.Biophys. Acta 113:51-56) slants or Petri dishes (5 cm diameter) 2-4 days at 37C. Spores were collected and resuspended in 1 ml of PBS-Tween 20 0.1%. Following centrifugation (5 min, 13,000 rpm), DNA was extracted from intact spores according to the manufacturer's instructions (Nucleon MiY kit, Amersham RPN 8518) except for a few details that are described below. Spores were first resuspended in 540 ul of Solution A. After addition of 60 ul of solution B, the mixture was vortexed for 30 sec and incubated 10 min at 70C. Then 300 ul of solution C were added and the mixture was homogenized by inverting the tube several times, allowed to cool for 5 min and subjected to a 5 min centrifugation at 13,000 rpm. The supernatant was collected and subjected to a second centrifugation (5 min, 13,000 rpm). DNA in the supernatant was precipitated with isopropanol, washed once with 70% ethanol, dried and resuspended in 50 ul solution D. Resuspension of the DNA was achieved by incubating the tubes 10 min at 65C. Typical yields were in the range of 2-3 ug DNA/prep. Data presented in Figure 1 show that the size of the DNA fragments averages 25 kb (Fig.1A) and that the DNA is suitable for further PCR analysis (Fig. 1B) or restriction enzyme analysis (Fig. 1C) and Southern hybridization (Fig.1D). Using this protocol, we have been able to prepare genomic DNA from up to 32 individual strains in less than 2.5 h with similar yields for most preparations.
The use of the Nucleon MiY kit to prepare genomic DNA from mycelium was also investigated. An A. fumigatus mat was surface-grown in 3 ml of liquid YEPD medium (0.3% Yeast Extract, 1% Bacto-peptone, 2% glucose) in a 5 cm diameter Petri dish for 24 h at 37C. Mycelium was collected with a sterile tooth-pick and allowed to dry on filter-paper. The mycelium mat was either directly incubated in Solution A or subjected to mechanical agitation for 30 sec in the presence of ca. 50 ul acid-washed 0.5 mm glass-beads and then resuspended in 540 ul Solution A. Genomic DNA was then extracted according to the procedure described above. DNA could only be detected in samples that had been subjected to mechanical disruption, suggesting that the Nucleon MiY kit is also suitable for the preparation of genomic DNA from mycelium provided that the mycelium has been previously homogenized.
The Nucleon MiY kit of Amersham provides a rapid, safe and convenient procedure for the preparation of a large number of samples of genomic DNA, a process that is required when analyzing transformants of Aspergillus sp. or when typing strains by RAPD or RFLP. It is of particular interest that the procedure is applicable to the preparation of genomic DNA from both intact spores and mechanically disrupted mycelium. Furthermore, the procedure is relatively inexpensive and cost-effective ($1.5/prep) It is likely to be useful for the preparation of genomic DNA from other fungal species.

Figure 1 : A. Ethidium bromide (EtBr)-stained agarose gel of three representative samples (lanes 1-3) of genomic DNA of A. fumigatus prepared from intact conidia using the Amersham Nucleon MiY kit. B. EtBr-stained agarose gel of PCR products obtained by amplification of 3 independent Nucleon MiY-purified A. fumigatus genomic DNA samples (lanes 1-3). Primer pairs were used that yield 150 bp and a 1.55 kb DNA fragments. C. EtBr-stained agarose gel of 3 independent Nucleon MiY-purified A. fumigatus genomic DNA samples digested with BamHI (lanes 1-3). D. Southern blot of the gel shown in C, probed with a 0.9 kb BamHI fragment of the A. fumigatus rho2 gene. Molecular weight markers (lambda DNA cut by HindIII in Panel A and 1kb Ladder in Panels B and C) are shown (lane m)