Isolation and cloning of the Sordaria macrospora ura3 gene and its heterologous expression in Aspergillus niger
Minou Nowrousian and Ulrich Kück - Lehrstuhl für Allgemeine Botanik, Ruhr-Universität Bochum, 44780 Bochum, Germany
We report the isolation and molecular characterization of the ura3 gene encoding orotidine monophosphate decarboxylase (OMPD) from Sordaria macrospora. Pulsed field gel electrophoresis and Southern hybridization revealed that the ura3 gene is located on chromosomes VI / VII of S. macrospora. A functional analysis of the S. macrospora ura3 gene was performed by DNA transformation using an Aspergillus niger pyrA mutant as recipient strain.
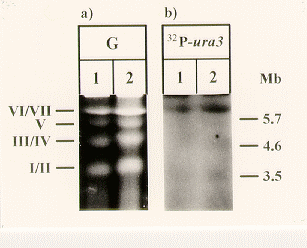
Southern hybridization of total DNA indicates that the ura3 gene is a single copy gene in S. macrospora (data not shown). As shown in Figure 2, chromosomes of two wild type isolates of S. macrospora were separated by pulsed field gel electrophoresis (Walz and Kück 1995 Curr. Genet. 29: 88-95) and probed with a DNA fragment carrying the ura3 gene. In both strains, the high molecular chromosomal band representing chromosomes VI and VII hybridizes with the ura3 gene specific probe.
In order to demonstrate the functionality of the ura3 gene, the uracil auxotrophic strain N593 of A. niger was transformed with plasmid p20.26. This plasmid has a size of 6.6 kb and contains the complete open reading frame of the S. macrospora ura3 gene with additional 5' and 3' nontranslated regions of 2.2 kb and 0.2 kb, respectively. The recipient strain A. niger N593 carries a mutation in the OMPD gene (pyrA, Goosen et al. 1987 Curr. Genet. 11: 499-503). It was transformed according to Mohr and Esser (1990 Appl. Microbiol. Biotechnol. 34: 63-70) with slight modifications as the transformants were selected for uracil prototrophy instead of antibiotic resistance. With a transformation rate of 14 transformants per g DNA, 55 uracil prototrophic transformants were isolated. The autoradiograph in Figure 3 shows the molecular analysis of six randomly selected transformants. In all of them one or more copies of the ura3 gene of S. macrospora are detectable, while the internal pyrA gene of the recipient strain is not labeled since hybridizations were performed under high stringency conditions. Our data prove that the isolated ura3 gene of S. macrospora is functional and can be used as a selection marker in transformation experiments.
We thank Dr. S. Pöggeler for providing the indexed cosmid library from S. macrospora and Prof. Dr. Tudzynski for the gift of the A. niger pyrA strain. This work was funded by the ´Deutsche Forschungsgemeinschaft´, Bonn - Bad Godesberg, Germany.
S.m. MSTTQ--QPHWSLQKSFAERVESSSHPLTSYLFRLMEVKQSNLCLSDDVEHARELLALADKIGP 62
N.c. MSTSQETQPHWSLKQSFAERVESSTHPLTSYLFRLMEVKQSNLCLSADVEHARELLALADKVGP 64
T.r. ------MAPHPTLKATFAARSETATHPLTAYLFKLMDLKASNLCLSADVPTARELLYLADKIGP 58
A.c. ------MAPHPTLKSTYSTRAETVTHPLSAYLYKLMDLKASNLCLSADVANARELLHVADKVGP 58
A.n. --------MSSKSQLTYTARASKHPNALAKRLFEIAEAKKTNVTVSADVTTTKELLDLADRLGP 56
S.c. -----------MSKATYKERAATHPSPVAAKLFNIMHEKQTNLCASLDVRTTKELLELVEALGP 53
A.t. SKNPTGKKLFDIMLKKETNLCLAADVGTAAELLDIADKVGP 41
B.t. GTHPVAAKLLRLMQKKETNLCLSADVSESRELLQLADALGS 41
* * * ** *** *
S.m. SIVVLKTHYDLITGWDYHPHTGTGAKLAALARKHGFLIFEDRKFVDIGSTVQKQYTAGTARIVE 126
N.c. SIVVLKTHYDLITGWDYHPHTGTGAKLAALARKHGFLIFEDRKFVDIGSTVQKQYTAGTARIVE 128
T.r. SIVVLKTHYDMVSGWTSHPETGTGAQLASLARKHGFLIFEDRKFGDIGHTVELQYTGGSARIID 122
A.c. SIVVLKTHYDMVAGWDFTPETGTGARLAKLARKHGFLIFEDRKFGDIGNTVELQYTQGAARIIE 122
A.n. YIAVIKTHIDILSDFSDETIEG----LKALAQKHNFLIFEDRKFIDIGNTVQKQYHRGTLRISE 116
S.c. KICLLKTHVDILTDFSM---EGTVKPLKALSAKYNFLLFEDRKFADIGNTVKLQYSAGVYRIAE 114
A.t. EICLLKTHVDILPDFT----PDFGSKLRAIADKHKFLIFEDRKFADIGNTVTMQYEGGIFKILE 101
B.t. RICLLKIHVDILNDFT----LDVMKELTTLAKRHEFLIFEDRKFADIGNTVKKQYEGGVFKIAS 101
* * * * * * ** ****** *** ** ** * *
S.m. WAHITNADIHAGEAMVSAMAQAAQKWRERIPYEVKTSVSVGTPVADQFADE------EAEDQVD 184
N.c. WAHITNADIHAGEAMVSAMAQAAQKWRERIPYEVKTSVSVGTPVADQFADE------EAEDQVE 186
T.r. WAHIVNVNMVPGKASVASLAQGAKRWLERYPCEVKTSVTVGTPTMDSFDDDADSRDAEPAGAVN 186
A.c. WAHIVNVNMVPGKASVTSLANAAAKWLERYPYEVKTSVTVGTPRNTDEDDD-DEDDGNAGEMER 185
A.n. WAHIINCSILPGEGIVEALAQTASAPDFSYGP-------------------------------- 148
S.c. WADITNAHGVVGPGIVSGLKQAA---EEVTKE-------------------------------- 143
A.t. WADIINAHVISGPGIVDGLKLKGMPR-------------------------------------- 127
B.t. WADLVNAHAVPGSGVVKVLEEVGLPL-------------------------------------- 127
** * * *
S.m. ELRKIVPRENSTSKEKDTDGRKGSIVSITTVTQTYEPADSPRLAKTISEGDEAVFPGIEEAPLD 248
N.c. ELRKVVTRETSTT-TKDTDGRKSSIVSITTVTQTYEPADSPRLVKTISEDDEMVFPGIEEAPLD 249
T.r. GMGSIGVLDKPIYSNRSGDGRKGSIVSITTVTQQYESVSSPRLTKAIAEGDESLFPGIEEAPLS 250
A.c. SHTFGLNGNNGVPIKESSDGRKGSIVSVTTVTQQYESAHSPRLTKTIAEEGDMLLAGLEEPPLN 249
A.n. ---------------------------------------------------------------E 149
S.c. ---------------------------------------------------------------P 144
A.t. ---------------------------------------------------------------G 128
B.t. ---------------------------------------------------------------H 128
S.m. RGLLILAQMSSKGCLMDGKYTWECVKAARKNKDFVMGYVAQQNLNGITKEDLAPGYEDGETSTE 312
N.c. RGLLILAQMSSKGCLMDGKYTWECVKAARKNKDFVMGYVAQQNLNGITKEALAPSYEDGESTTE 317
T.r. RGLLILAQMSSQGNFMNKEYTQASVEAAREHKDFVMGFISQETLN------------------- 295
A.c. RGLLILAQMSSAGNFMNAEYTQACVEAAREHKDFVMGFVSQEALN------------------- 294
A.n. RGLLILAEMTSKGSLATGQYTTSSVDYARKYKNFVMGFVSTRSLGEVQSEVSS----------- 202
S.c. RGLLMLAELSCKGSLATGEYTKGTVDIAKSDLDFVIGFIAQRDMGG------------------ 190
A.t. RGLLLLAEMSSAGNLATGDYTAAAVKIADAHSDFVMGFISVNPASWKC---------------- 176
B.t. RACLLVAEMSSAGTLATGSYTEAAVQMAEEHSEFVIGFISGSRVSMK----------------- 175
* * * * ** * * ** *
S.m. EEAQADNFIHMTPGCKLPPPGEEAPQ-----GDGLGQQYNTPDNLVNIKGTDIAIVGRGIITAS 371
N.c. EEAQADNFIHMTPGCKLPPPGEEAPQ-----GDGLGQQYNTPDNLVNIKGTDIAIVGRGIITAA 372
T.r. -TEPDDAFIHMTPGCQLPPEDEDQQTNGSVGGDGQGQQYNTPHKLIGIAGSDIAIVGRGIIKAS 358
A.c. -SQPDDDFIHMTPGCQLPPEHEEDAE---LRGDGKGQQYNTPEKLIGVCGADIVIVGRGIIKAG 354
A.n. -PSDEEDFVVFTTGVNISSK-----------GDKLGQQYQTPASAIG-RGADFIIAGRGIYAAP 253
S.c. -RDGEYDWLIMTPGVGLDDK-----------GDALGQQYRTVDDVVS-TGSDIIIVGRGLFAKG 241
A.t. -GYVYPSMIHATPGVQMVKG-----------GDALGQQYNTPHSVITERGSDIIIVGRGIIKAE 228
B.t. -----PEFLHLTPGVQLEAG-----------GDNLGQQYHSPQEVIGKRGSDIIIVGRGIIASA 223
* * ** **** * * * ***
S.m. -DPPAEAERYRRKAWKAYQDRRERLA 396 S.m. = Sordaria macrospora (Z70291)
N.c. -DPPAEAERYRRKAWKAYQDRRERLA 397 N.c. = Neurospora crassa (M13448)
T.r. -DPVEEAERYRSAAWKAY---TERLLR 381 T.r. = Trichoderma reesei (S14132)
A.c. -DLQHEAERYRSAAWKAY---TER-VR 376 A.c. = Acremonium chrysogenum (P14017)
A.n. -DPVQAAQQYQKEGWEAYLARVGGN 277 A.n. = Aspergillus niger (P07817)
S.c. RDAKVEGERYRKAGWEAYLRRCGQQN 267 S.c. = Saccharomyces cerevisiae (P03962)
A.t. -NPAETAHEYRVQGWEAYLEKCSQ 251 A.t. = Arabidopsis thaliana (Q42586)
B.t. -NQLEAAKMYRKAAWEAYLSRLAV 246 B.t. = Bos tauris (P31754)
* * **
Figure 1.
Comparison of amino acid sequences from OMPD genes from different eukaryotes. The sources and corresponding accession numbers (EMBL data library) are given at the end of all sequences. In the cases of Arabidopsis thaliana and Bos tauris, only the OMPD domains of the polypeptides were used. Asterisks below the sequence indicate amino acid residues, which are conserved in all sequences. Dashes represent gaps introduced to obtain the best alignment.

Figure 2. Chromosomal mapping of the ura3 gene from S. macrospora. a) Chromosomes of two S. macrospora wild type strains (isolate S1957, lane 1; isolate L3346, lane 2) were separated using pulsed field gel electrophoresis. Roman numerals indicate positions of S. macrospora chromosomes. On the right, sizes of marker chromosomes are given in Mb. b) Autoradiograph showing the chromosomal localisation of the ura3 gene. The filter bound chromosomal DNA was probed with a DNA fragment representing the open reading frame and adjacent regions of the ura3 gene.
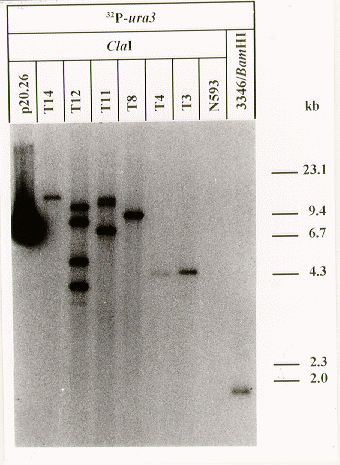
Figure 3. Autoradiograph from a Southern hybridization experiment with DNA from A. niger transformants. DNA from six transformants (T3, T4, T8, T11, T12 and T14) and from the recipient strain N593 was digested with ClaI and separated by gel electrophoresis. The Southern blot was probed with a DNA fragment, representing the open reading frame and adjacent regions of the ura3 gene. As a control, the transforming plasmid p20.26 and DNA from a S. macrospora wild type strain (L3346) were used. Sizes of marker fragments are given on the right margin.