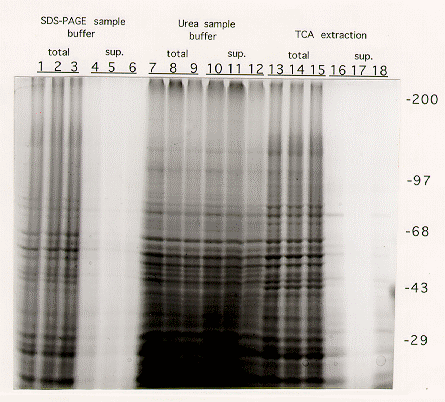
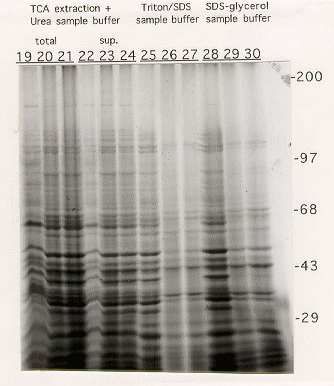
Optimization of protein extraction from Aspergillus nidulans for gel electrophoresis
Nir Osherov and Gregory S. May- Department of Cell Biology, Baylor College of Medicine, Houston, Texas, 77030 USA.
Efficient extraction of proteins from filamentous fungi is difficult both as a result of high endogenous protease activity, the existence of a hard cell wall, and the tendency of hyphae to clump. We have had difficulty using published protocols in extracting high molecular weight proteins (>150 kDa), and in extraction from germlings grown on minimal media with a poor carbon source such as glycerol. In order to address these problems, we have compared several different extraction protocols and found the protocol utilizing 9M Urea-sample buffer to be the fastest and most efficient with sample variation being kept to a minimum.
Aspergillus nidulans (strains derived from GR5) conidia were germinated in rich liquid medium (0.5% yeast extract, 1% dextrose), or glycerol minimal medium for 14 and 24 hours respectively. Germlings were scraped off the plates, washed once in distilled water and dried on paper towels. Cells were then frozen in liquid nitrogen, and lyophilized overnight. The lyophilized pellets were ground up in a mortar and pestle weighed at 10 mg dry weight per extraction reaction and an equal volume of glass beads was added.
The following extraction procedures were then performed:
SDS-PAGE sample buffer extraction: 0.2 ml of sample buffer containing 1% SDS, 10% glycerol, 25 mM Tris-HCl pH 6.8, 1 mM EDTA, 0.7 M mercaptoethanol, was vigorously mixed with the lyophilized powder, boiled for 2 min, vortexed for 1 min, then boiled for another minute.
Urea sample buffer extraction: 0.2 ml of sample buffer containing 1% SDS, 9 M urea, 25 mM Tris-HCl pH 6.8, 1 mM EDTA , 0.7 M mercapto-ethanol was vigorously mixed with the lyophilized powder, boiled for 2 min, vortexed for 1 min, then boiled for another minute.
TCA extraction: 0.5 ml of 10% TCA was mixed and vortexed with the lyophilized powder, incubated 5 min at room temperature, then washed 3 times in 90% acetone, 20 mM HCl, and air dried. An equal volume of glass beads was added together with 0.2 ml of SDS-PAGE sample buffer, mixed and then boiled for 2 min, vortexed for 1 min, and boiled for another minute.
TCA and urea sample buffer extraction: Lyophilized powder was treated as in protocol (3), followed by extraction of the TCA precipitate in 9 M urea sample buffer as described in (2).
Triton X100/SDS buffer extraction: 0.1 ml buffer containing 50 mM Tris-HCl pH 7.5, 200 mM NaCl, 5 mM EDTA, 0.25% Triton X100, 0.1% SDS, 1 mM PMSF, aprotinin and leupeptin at 10 micrograms/ml was vigorously mixed with the lyophilized powder. The cellular debris was spun down in a microcentrifuge for 5 min at maximal speed, and the supernatant was mixed with 2X SDS-PAGE sample buffer and boiled for 3 min.
SDS-glycerol buffer extraction: 0.1 ml buffer containing 200 mM Tris-HCl pH 7.5, 25% glycerol, 1% SDS and 1 mM PMSF was vigorously mixed with the lyophilized powder. The cellular debris was spun down in a microcentrifuge, and the supernatant was mixed with 2X SDS-PAGE sample buffer and boiled for 3 min.
Total mycelial extracts, containing both soluble and insoluble material, or the supernatant collected after microcentrifugation were then separated on a 6% SDS-PAGE gel. Proteins were visualized by staining in Coomassie Blue.
Results: Extraction procedures performed with SDS-PAGE sample buffer(1) or TCA precipitation followed by sample buffer(3) failed to extract proteins efficiently from the cellular debris (Figure 1: compare lanes 1-3 to 4-6, and 13-15 to 16-18). After centrifugation most of the proteins remained in the pellet. Furthermore, proteins from total cell extracts (debris mixed with sample buffer) ran as blurry bands, particularly at high molecular weights, probably as a result of contamination by high molecular weight DNA (see lanes 1-3 and 13-15).
Extraction procedures using either Triton-X100/SDS (5) or SDS-glycerol (6) buffers resulted in very inefficient extraction of high molecular weight proteins from cells grown on glycerol (Figure 1, lanes 26,27,29,30).
The best results were obtained using either the urea sample buffer(2) or TCA/urea sample buffer (4) extraction protocols (Figure 1, lanes 7-9/10-12 and 19-21/22-24). In both cases, proteins of all sizes were efficiently extracted into the supernatant. No major difference was seen in the ability to extract protein from cells grown on either YAG or glycerol. The TCA/urea protocol, though slightly longer, gave a slightly better protein yield, and the protein bands appeared to be more discrete.
For efficient extraction of both high and low molecular weight proteins, irrespective of growth conditions or strain, in which sample variation is kept to a minimum, we therefore recommend using either protocol (2) or (4).


Figure 1. SDS-PAGE analysis of Aspergillus nidulans proteins extracted as described in the text.
The gels shown contain 6% acrylamide. After fixation, gels were stained with Coomasie blue for protein visualization. Extracts run on the first lane of each triplet were derived from mycelia grown on YAG rich medium, whereas those on the second and third lanes were from mycelia grown on glycerol minimal medium.