Isolation by genomic subtraction of subspecies-specific DNA probes from Verticillium dahliae
N.A. Patterson* and K.F. Dobinson - Agriculture and Agri-Food Canada, 1391 Sandford St., London, ON Canada N5V 4T3
*present address: Plant Biotechnology Centre, Department of Biological Sciences, University of Calgary, Calgary, AB Canada T2N 1N4.
The vascular wilt pathogen Verticillium dahliae Kleb. has a very broad host range, including more than 200 different plant species in 45 families. Most isolates cannot be discriminated morphologically, and reliable, rapid methods for classifying and differentiating strains are needed. We have used a genomic subtraction method to isolate DNA probes that can be used to differentiate V. dahliae isolates and to investigate the genetic variation within this fungal species.
The V. dahliae isolates used for genomic subtraction experiments included Dvd-2 (vegetative compatibility group (VCG) 4B, race 2 tomato pathogen), Dvd-T5 (VCG 2A, race 1 tomato pathogen), and Dvd-E6 (VCG 4A, tomato nonpathogenic). These isolates were chosen on the basis of molecular differences previously identified by randomly amplified polymorphic DNA (RAPD) fingerprinting and rDNA RFLP analysis (Dobinson et al. Mycol. Res. in press), with the objective of isolating DNA sequences specific to the race 1 and/or 2 V. dahliae isolates.
The basis of the genomic subtraction method, originally developed by Straus and Ausubel (1990 Proc. Natl. Acad. Sci. USA 87:1889-1893), is the enrichment of DNA sequences that are present in one isolate (designated the "target"), and absent in another isolate (the "driver"). The method involves the sequential hybridization of an excess of biotinylated driver DNA with a small amount of target DNA, and removal with streptavidin, of driver DNA-target DNA hybrids and driver DNA. Following several cycles of subtraction, the enriched target DNA sequences can be amplified by PCR, and cloned into a plasmid vector. The subtraction procedure used in this study was essentially that described by Straus and Ausubel (1990), except that the biotin acetate was photoactivated by illumination for 30 min with a 150 W GE Reflector Floodlamp, and during each subtraction cycle, Streptavidin Magnesphere® Paramagnetic Particles (Promega Corp.) were used to remove biotinylated driver DNA, and driver DNA-target DNA hybrids. Six rounds of subtraction were done, using either Dvd-2 DNA as the target and Dvd-E6 DNA as the driver, or a hygromycin-resistant transformant of Dvd-T5 (VDT8C22) as the target and Dvd-2 as the driver. Total subtraction products were amplified by PCR using the primers and conditions described by Straus and Ausubel (1990). 32P-radiolabeled amplification products were hybridized to Southern blots of genomic DNA from Dvd-2, Dvd-T5 and Dvd-E6, to confirm the removal of sequences common to both driver and target DNA (data not shown).
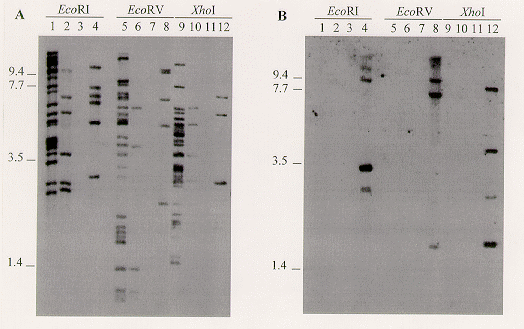
In the case of the Dvd-2/Dvd-E6 subtractions, the amplified subtraction products were digested with Sau3AI and cloned into the BamHI site of pBluescript® II KS+ (Stratagene). Individual clones were selected at random and screened by Southern blot hybridization analysis. The 310 bp sequence (designated E18) in clone pBNKE18 was present in low to moderate copy number in the race 2 tomato pathogens Dvd-2, Dvd-S85 (VCG 2A), and Dvd-3 (VCG 4B), but absent from Dvd-E6 (Figure 1A). Hybridization to electrophoretically separated chromosomes indicated that this sequence is distributed throughout the V. dahliae genome (data not shown). The race 1 isolate Dvd-T5 and the race 2 isolate Dvd-S85 share the same E18 banding pattern (Dobinson et al. Mycol. Res. in press), suggesting that although these isolates differ in cultivar specificity, they are closely related, and indicating that the E18 sequence is not specific to the race 2 pathotype.
The products obtained from the VDT8C22/Dvd-2 subtraction were amplified and labeled with 32P, and used to screen a Dvd-T5 genomic DNA library constructed in GEM11. Recombinant clones that hybridized to the probe were further characterized, and restriction fragments that hybridized with the subtraction product probe were subcloned into pBluescript® II KS+. Clone pBGC22-S contains a 4.4 kb SacI restriction fragment isolated by this method. This sequence (BGC22-S) was present, at low copy number, only in the VCG 2A isolates Dvd-S85 and Dvd-T5, further indication of the relatedness of these two isolates (Figure 1B and data not shown).
In summary, using genomic subtraction we have isolated from V. dahliae two subspecies-specific repetitive DNA sequences, that can differentiate V. dahliae strains, and should therefore be useful for evaluating genetic diversity within the species. Experiments are also in progress to assess the association of the sequences with specific vegetative compatibility groups and other biological characters.

Figure 1. Southern blot analysis of V. dahliae genomic DNA, digested with EcoRI, EcoRV or XhoI, and hybridized with the E18 probe (panel A), then stripped of the probe and hybridized with the BGC22-S probe (panel B). High stringency hybridization and wash conditions were as previously described (Dobinson et al. Mycol. Res. in press). Probes were generated by PCR amplification from the recombinant plasmids, using a PCR DIG DNA labeling kit (Boehringer Mannheim) and primers homologous to the vector T7 and T3 promoter sequences flanking the cloning site. Lanes 1, 5 and 9, Dvd-2; lanes 2, 6 and 10, Dvd-3; lanes 3, 7 and 11, Dvd-E6; lanes 4, 8 and 12, Dvd-S85. Positions of molecular weight standards (kb) are indicated to the left of each panel. Ethidium bromide staining of the gel indicated that equivalent amounts of DNA were applied to each lane (not shown).