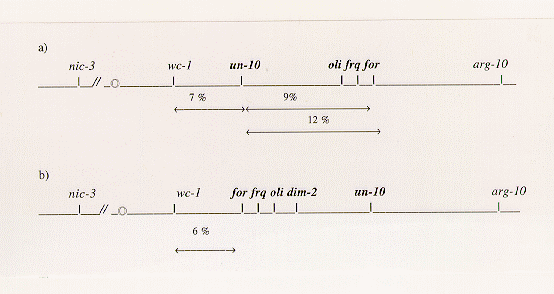Inversion in the published genetic map of linkage group VII
Elena
Kouzminova and Eric U. Selker* University of Oregon, Institute of Molecular Biology Eugene,
Oregon
*corresponding author
Fungal Genetics Newsletter 46:14-15
In the course of cloning the dim-2 gene of Neurospora crassa we
found that the published map of LG VII has an inversion of a segment extending from for
to un-10. Direct physical mapping confirmed that the gene order in this
region should be wc-1, for, frq, oli, un-10.
In preparation to clone the dim-2 gene (Foss et al. 1993 Science
262:1737-1741) of Neurospora crassa, we mapped this gene to the
right arm of linkage group VII (LGVII) between wc-1 and arg-10. To
map the dim-2 gene more precisely, three other crosses were performed (Table 1).
We used strains with genetic markers wc-1, frq and for on
LGVII and initially relied on a published genetic map (Figure 1a) to interpret our results. Data
from the cross between un-10 and wc-1 dim-2 strains suggested that
dim-2 is left of un-10 and between wc-1 and
un-10; about half of the recombinants (21/36) in the wc-1 to un-10
interval were dim-2 (Table 1, cross 1). In two additional crosses in which one
of the parents carried either a for or a frq mutation, however, we could
not find dim-2 recombinant progeny of the genotype wc-1
dim-2 for and wc-1 dim-2 frq
(Table 1, crosses 2 and 3); other markers in these crosses segregated as expected from the
published genetic map (data not shown). Therefore, dim-2 appeared to be tightly
linked to both for and frq. This result was unexpected since frq
and for were reported to be to the right of un-10 by
approximately 9 and 12 map units, respectively (Figure 1a). To explain our genetic data
we considered the possibility that the segment including un-10, oli, frq
and for is inverted (Figure 1b). This orientation accounted for all our data and
was subsequently confirmed by physical mapping (data not shown). Results of chromosomal
walks from wc-1 and un-10 showed that for is proximal to
wc-1 followed by frq, oli, dim-2 and
un-10 (Kouzminova and Selker, unpublished). Thus we suggest that the order of the
genes for, frq, oli and un-10 is as shown in
Figure 1b.
Table 1. Map data.
|
Cross |
Zygote genotype
|
Genotype and number of
recombinant progeny analyzed
for dim-2 |
Genotype and number of dim-2 recombinant progeny |
|
1 a |
+ + un-10
wc-1 dim-2 + |
wc-1+ un-10+
36 |
wc-1+ dim-2+ un-10+
21
wc-1+ dim-2 un-10+
15 |
|
2 b |
+ for (+)
wc-1 + (dim-2)d |
random progeny
100 |
wc-1 for dim-2+ 4
wc-1+ for+ dim-2
2 |
|
3 c |
+ frq::hph (+)
wc-1 + (dim-2)d |
wc-1 frq::hph
10 |
wc-1 frq::hph dim-2+
10
wc-1 frq::hph dim-2 0 |
aRecombinant progeny were selected on minimal medium at 34oC to
select against un-10 progeny. The wc-1 mutation
was scored under constant light at 34oC.
b Progeny were scored without selection.
c Progeny were selected on plates supplemented with hygromycin and scored for
wc-1 under constant light at 34oC.
d Parentheses indicate that the position of dim-2 relative to either
for or frq could not be determined from the cross.
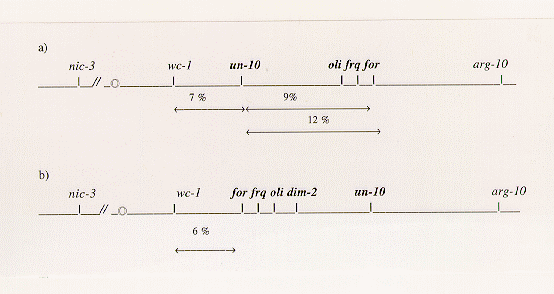
Figure 1. (a) Previous map and percentages of recombination between some genetic markers
on LGVII (Perkins et al. 1982 Microbiol. Reviews, 46:426-570). (b) Revised genetic
map based on our findings. Genetic distance between wc-1 and for was
calculated from the cross 2 data (Table 1). the centromeric region is indicated by the circle on the
left and genes in the disputed region are in bold.
Return to the FGN 46 Table of Contents
Return to the FGSC main page