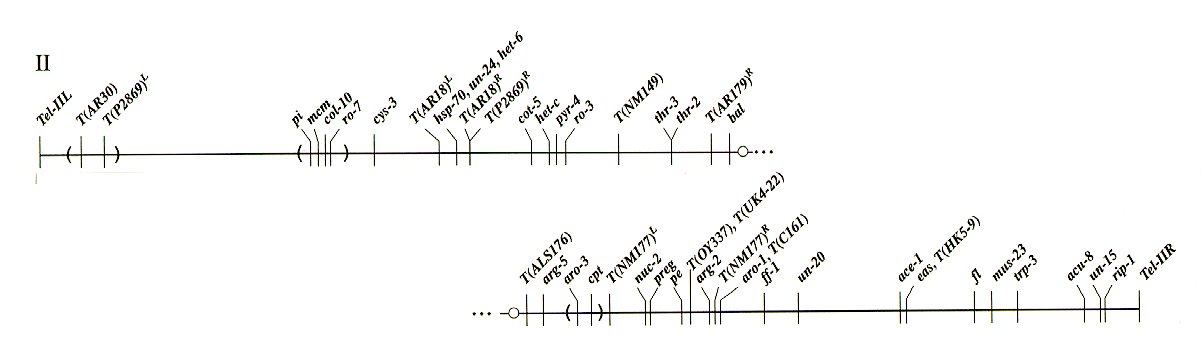
LOCI IN LINKAGE GROUP II
aag-1 Linked to acu-5 (26%), arg-5 (33%)
ace-1 Between un-20 (15%) and eas (1%),
fl (11%)
ace-9 Between nuc-2 (2%) and arg-12 (3%).
acr-5 Between arg-5 (6%) and pe (9%).
acu-5 Linked to arg-5 (6%), aro-3 (7%).
acu-8 Between trp-3 (8%) and un-15 (3%).
acu-12 Right of Cen-II. Linked to trp-3.
acu-13 Linked to trp-3 (23%).
alc-1 Linked to pe (10%).
aod-2 Linked to arg-5 (7%), thr-3 (16%).
arg-5 Between bal (1%; 9%), T(ALS176) and
aro-3, pe (6%, 18%).
arg-12 Between pe (1%; 5%) and
T(NM177)R, aro-1 (<1%).
aro-1 Between arg-12 (<1%),
T(NM177)R and ff-1 (4%; 6%).
aro-3 Between arg-5 (1%; 3 %) and
T(NM177)L, nuc-2.
arp3 Near arg-12.
atp-2 Linked to Cen-II, arg-5 (0T/18 asci).
bal Between T(AR179) and
T(ALS176), arg-5 (1%; 7 %). Probably left of
Cen-II.
bli-4 Linked to Fsr-34 (0T/18 asci). Between
eas (1T/18 asci) and leu-6 (9T, 1NDP/18 asci).
cdr-1 Left of arg-12 (35%).
Cen-II Between T(AR179)R
and T(ALS176), arg-5 (0T/18 asci).
cit-1 Linked to preg (0T/18 asci).
col-10 Left of cys-3 (14%). At or near pi.
con-6 Left of arg-12 (2 or 3/18).
cot-5 Between T(P2869), T(AR18)R
and het-c (3%).
cpd-4 Right of arg-12 (7%).
cpt Between arg-5 (3%) and
T(NM177)L, pe (6%).
cwl-1 Between arg-5 (3%; 6%) and arg-12
(12%).
cwl-2 Between arg-12 (10%), un-20 (4%) and
fl (9%).
cya-4 Between arg-5 (2/39; 1T/18 asci) and
preg (1T + 1 double exchange/18 asci).
cyb-3 Between Tip-IIL and ro-3 (9%).
cyc-1 Between thr-3 (38%) and trp-3 (18%).
cys-3 Between ro-7, pi (4%) and
T(AR18)L .
da Linked to thr-3 (3%); right of T(NM149).
dim-4 Linked to arg-12, aro-1,
ure-3.
eas Between ace-1 (1%) and fl (9%).
en(am)-2 Linked to pe.
ff-1 Between aro-1 (5%) and un-20 (4%).
fl Between ace-1, eas (9%) and
mus-23 (1%; 2%), trp-3 (3%).
fs-2 Probably linked to cot-5 (14/48).
Fsp-1 Right of pe (4%).
Fsr-3 Between vma-2 (1T/18 asci) and eas
(1T/18 asci).
Fsr-17 Between trp-3 (1/16) and leu-6 (0/16;
9T, 1NPD/18 asci).
Fsr-21 Between arg-12 (1/17) and trp-3 (1/17;
4/41).
Fsr-32 Between pyr-4 (10/41) and Cen-II
(1T/18 asci), arg-5 (2/41).
Fsr-34 Between arg-12 (1/18), eas (1T/18 asci)
and trp-3 (1/17). Linked to Fsr-21 (0/59).
Fsr-52 Between pyr-4 (2/41) and Cen-II
(5T/18 asci), arg-5 (0/41)
Fsr-54 Between pyr-4 (2/41), Fsr-52 (3/18, 8
41) and arg-5 (10/41).
Fsr-55 Between arg-5 (1T/18 asci; 2/39) and preg
(4/39). Linked to cya-4 (0T/18 asci).
glp-2 Between T(ALS176). arg-5 ((8%) and
T(NM177)L, pe (7%).
gpd-1 Linked to arg-12 (0/18), vma-2 (1T/18
asci), eas (3T/18 asci).
hbs Left of con-6 (1 or 2/18), arg-12 (3 or
4/18).
het-c Between cot-5 (3%) and Pad-1,
pyr-4 (1%), ro-3.
het-d Right of fl (25%).
het-6 Between cys-3,
T(AR18)L, cys-3,
hsp70, un-24 and
T(AR18)R, T(2869),
cot-5.
hH3 Between Fsr-55 and Fsr-3. On same
plasmid with aro-9.
hH4-1 Adjoins
hH3.hsp70 Between
T(AR18)L and un-24 .
leu-6 Between trp-3 (2/16), Fsr-17 (5T/18
asci), and Tel-IIR (1T, 1NPD/16asci).
lp Right of thr-3 (10%).
mcm Linked to ro-7 (1%).
mus-7 Between arg-5 (8%; 12%) and nuc-2
(11%).
mus-23 Between fl (<1%; 2%) and trp-3
(9%).
mus-24 Left of pyr-4 (8%).
mus-27 Right of nuc-2 (16%).
nik-2 Linked to arg-12 (0/17).
nrc-2 Linked to cya-4, Fsr-55.
nuc-2 Between aro-3,
T(NM177)L and preg (1 or 2%),
pe (4%).
nuc-3 Right of arg-12 (4%). (nuc-4 -
nuc-7?)
nuh-5 Linked to trp-3 (30%), T(4637).
nuh-9 Linked to arg-5. Left of mus-7,
nuc-2.
nuo78 Linked to preg (0T/5 asci).
osr-2 Linked to arg-5 (£12%).
Pad-1 Between het-c and pyr-4 (1%).
pe Between nuc-2 (4%) and arg-12 (1%, 5%).
pi Linked to ro-7 (0/75). Between T(AR30)
(28%) and cys-3 (4%).
pma-1 Between cya-4 (2T/18 asci) and preg
(1T/15 asci).
pmg Linked to pyr-4 (<1%).
prd-5 Between arg-12 (4%; 10%) and un-20
(8%).
preg Between T(NM177)L,
nuc-2 (1 or 2 %) and arg-12,
T(NM177)R.
pyr-4 Between het-c (1%), Pad-1 (1%) and
ro-3 (1 or 2 %).
rap-1 Adjacent to vma-6. Near Cen-II.
rip-1 Between un-15 (1%) and Tip-IIR.
ro-3 Between pyr-4 (1 or 2 %) and
T(NM149), thr-2 (6%; 25%).
ro-7 Linked to pi (0/75). Left of cys-3
(11%).
scr Linked to arg-12 (2%), probably to the left.
spco-14 Linked to arg-5 (7%).
su([mi-1])-3 Linked to fl (31%).
su([mi-1])-4 Linked to fl (22%),
arg-5 (40%).
su([mi-1])-10 Linked to arg-5
(29%), fl (24%).
T(UK4-22) Linked to pe (0/46) and arg-12,
aro-1 (0/23).
T(AR18)L Between ro-7,
cys-3 and hsp70.
T(AR18)R Between het-6
and cot-5. Within 5.6 kb of T(P2869).
T(AR30) Between Tip-IIL and pi (28%).
T(NM149) Between ro-3 and thr-2, thr-3.
T(ALS176) Between Cen-II and arg-5.
T(NM177)L Between aro-3
cpt and nuc-2.
T(NM177)R Between arg-12 and
aro-1.
T(OY337) Between pe and arg-12.
T(P2869)L Between Tip-IIL and
ro-7.
T(P2869)R Between het-6 and
cot-5. Within 5.6 kb of T(AR18)R.
Tel-IIL Linked to hsp70 (2T/18 asci).
Tel-IIR Right of leu-6 (1T, 1NPD/18 asci).
Tip-IIL Left of T(AR30), pi (³28%).
Tip-IIR Right of un-15, rip-1.
thr-2 Between ro-3, T(NM149) and
T(AR179), bal, arg-5 (3%; 18%).
thr-3 Linked to thr-2 (<0.1%).
tng Between pyr-4 (16%), T(NM149) and
arg-5 (6%; 14%). Linked to thr-2 (2%).
trp-3 Between fl (2%; 6%), mus-23 (6%) and
un-15 (10%).
tu Between pe (8%) and fl (19%).
uc-1 Linked to pyr-4 (31%).
un-15 Between trp-3 (10%) and rip-1 (1%).
un-20 Between aro-1 (5%; 9%), ff-1 (4%)
and ace-1 (15%).
un-24 Between T(AR18)L,
hsp70 and het-6,
T(AR18)R.
ure-3 Between arg-12 (7%; 12%) and un-20
(8%).
vma-2 Between preg (6T/18 asci) and Fsr-3
(1T/18 asci), eas (2T/18 asci).
vma-6 Near Cen-II. In same cDNA fragment with
rap-1.
vma-10 Linked near arg-12.
vph-1 Left of Cen-II (5T/18 asci), Fsr-32
(4T/18 asci). Linked to Fsr-52 (0T/18 asci).
xdh-1 Linked to pe (14%).
ypt-1 Linked to eas (0T/17 asci).