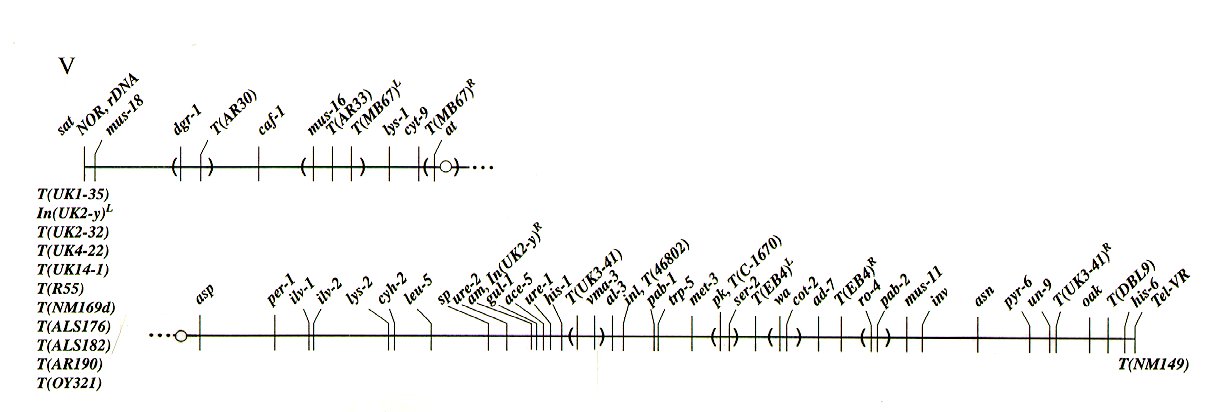
LOCI IN LINKAGE GROUP V
ace-5 Between gul-1 (<1%) and ure-1
(<1%).
act Linked near inl.
acu-1 Right of asn (21%).
acu-3 Between inl (7%) and asn (20%) .
ad-7 Between cot-2 (4%) and
T(EB4)R, ro-4 (4%),
pab-2 (8%).
al-3 Between pho-2 and inl (1%; 1T/18
asci).
am Between ure-2 (2%) and gul-1
(<1%), ace-5 (<1%), ure-1 (1%; 150kb).
arg-4 Between sp (1%; 11%) and inl (2%;
4%).
Asc(KH2A83) Left of al-3; probably right of
his-1.
Asm-1 Linked to al-3 (0T/18 asci).
asn Between inv (4%; 9%) and pyr-6 (6%).
asp Between at (0%; 3%) and per-1 (16%;
26%).
at Linked to Cen-V (0/57 asci). Between cyt-9
(2%; 5%) and asp (0%; 3%) .
caf-1 Between dgr-1 (8%), T(AR30) (19%)
and mus-16 (4%), T(AR33) (5%; 12%).
ccg-1 Between Cen-V (4T/18 asci) and ilv-2
(4T/18 asci).
ccg-6 Linked to al-3, inl (0 or 1T/18 asci).
ccg-8 Linked to lys-1, Cen-V (0T/18 asci).
Cen-V Between lys-1 (5%), cyt-9 (2%),
T(MB67)R and asp.
Linked to at (0T/57 asci).
chol-3 Between at (18%) and al-3 (19%;
47%).
chs-1 Between con-4a, Fsr-9 (1/18) and am
(4/18).
chs-3 Between am (1 or 2/18) and inl (1 or
2/18).
cit-2 Linked to inl (1T/18 asci).
cl Right of pk (1 or 2%).
cmd Between inl (3T/18 asci), cya-2 (2T/18
asci) and tom70 (1 or 2T/17 asci).
cna-1 Linked to cyh-2.
col-9 Between inl (16%) and asn (5%).
con-2 Linked to rDNA (2/18; 4T/18 asci) and con-4a
(2/18), lys-1 (8T/18 asci).
con-4a Between rDNA (4/18), con-2 (2/18)
and chs-1 (1 or 2/18), am (4/18).
con-4b On same plasmid as con-4a.
cot-2 Between ser-2 (5%),
T(EB4)L and ad-7 (4%).
cot-4 Between rol-3 (5%) and inl (10%).
cpc-3 Linked to cyh-2 (0 or 1/16).
crp-4 Right of inl (9T/18 asci), Fsr-20 (1T/18
asci). Linked to sdv-6 (0T/18 asci).
cya-2 Linked to al-3 (3%), inl (0T/18 asci).
cya-7 Right of Cen-V.
cyb-1 Between al-3 (24%) and his-6 (10%).
cyh-2 Between lys-2 (<1%) and leu-5 (1 or
2%).
cyt-9 Between lys-1 (5%) and
T(MB67)R, Cen-V (4T/57 asci),
at (5%; 2T/55 asci).
dgr-1 Between T(UK2-33),
In(UK2-y)L and T(AR30),
caf-1 (7%).
dim-3 Between am and inl.
en(am)-1 Between am (8%) and inl (1%).
erg-1 Between pk (2%) and asn (9%).
erg-2 Left of inl (6%).
esr-1 Between pyr-6 and inl (7%).
fdh Contiguous with leu-5.
fkr-1 Between leu-5 (4%) and ure-2 (13%).
fkr-2 Between inl (10%) and cot-2 (3%).
fpr-1 Linked to cyh-2 (<1%).
fpr-4 Right of inl (11%).
Fsr-9 Linked to Cen-V, lys-1 (1T/18 asci),
con-4, Fsr-16 (0T/18 asci).
Fsr-16 Linked to Cen-V, lys-1 (1T/14 asci).
Fsr-20 Right of ro-4 (1/18).
gln-1 Linked to inl (2%), probably to the right.
glp-6 Left of inl (30%).
gna-2 Linked to inl (0/18).
gran Between pab-2 (1%; 8%) and his-6
(8%; 27%).
gul-1 Between am (<0.1%) and ace-5
(<1%).
hah Linked to inl (13%).
hgu-4 Between cyh-2 (7%) and ure-2 (10%).
hH1 Linked to ccg-9, pho-4.
his-1 Between am (3%), ure-1 (1%) and
pho-2 (3%), al-3, inl (1%; 10%).
his-6 Between oak (6%) and T(NM149)
(0/499), Tel-VR (30 kb).
hsp80 Between inl (3T/18 asci), cya-2 (2T/18
asci and tom70 (1/17 asci).
hsp83 Linked to cmd, hsp80 (0/18 asci).
Iasc Right of Cen-V.
ilv-1 Between per-1 (4%) and ilv-2 (<1%;
9%), lys-2 (4%; 7%).
ilv-2 Between ilv-1 (<1%; 9%) and lys-2.
In(UK2-y)L Breakpoint in NOR.
In(UK2-y)R am Breakpoint at am.
inl Between al-3 (1%; 1/18 asci) and pab-2
(1%;10%).
inv Between ro-4 (5%; 8%), pab-2 (3%),
mus-11 (4%) and asn (4%; 9%).
leu-5 Between cyh-2 (1%) and sp (3%; 9%).
lis-3 Right of inl (4%).
lys-1 Between caf-1 (4%; 7%) and cyt-9
(5%), at (1%; 7%; 20%).
lys-2 Between ilv-1, ilv-2 (4%; 7%) and
cyh-2 (<1%), leu-5 (9%).
mc Linked to cyh-2.
mcb Linked to al-3 (3%; 1T/18 asci), cyh-2,
sod-2 (0T/18 asci).
md Between sh (3%) and sp (9%).
mei-2 Between inv (18%) and his-6 (12%).
met-3 Between pab-1 (1%), trp-5 (4%) and
pk (1%).
mfa-1 Linked to leu-5 (0T/18 asci).
mus-11 Between ad-7 (6%; 11%), pab-2 (7%)
and inv (4%), un-9 (10%), mei-2 (5%; 10%).
mus-12 Left of inl (10%).
mus-16 Between caf-1 (4%) and lys-1 (9%).
mus-18 Linked to rDNA (0T/17 asci), con-2
(2/17 asci), T(AR190) (£3%).
mus-28 Left of lys-1 (10%).
mus-34 Between leu-5, ure-2 and
am, his-1 (3%).
mus-37 Between lys-1 and cyh-2.
mus(SCl7) Left of inl (27%).
nap Linked to inl (15%). Right of ure-2
(32%).
ndc-1 Left of arg-4 (2%), inl (6%).
nmr Between am (3%; 7%) and gln-1 (4%;
10%).
NOR Between Tip-VL, sat and
T(AR30), dgr-1. Involved in numerous rearrangements.
nuh-3 Between cyh-2 (4%) and al-3 (17%).
nuo21.3a Left of inl (18%), inv (22%) .
nuo24 Linked to cyh-2 (0T/18 asci).
oak Between un-9 (7%) and
T(DBL9), his-6 (6%).
pab-1 Between inl (1%; 10%) and trp-5,
met-3 (1 or 2%).
pab-2 Linked to ro-4 (0/407). Between ad-7
(8%), T(EB4)R and inv (3%).
pcna Linked to rDNA (0T/18 asci).
per-1 Between asp (16%; 26%) and ilv-1
(4%).
pho-2 Between his-1 (3%) and inl (4%).
pk Between met-3 (1%) and
T(EB4)L, cot-2 (8%).
pl Linked to gran (0/75). Between asn (1%; 9%)
and his-6 (16%).
Pogo Adjoins Tel-VR.
poi-1 Linked to cyh-2 (0T/18 asci).
pold Between ure-2 (<70kb) and am
(10kb).
ppt-1 Linked to inl (0/18).
prd-2 Between lys-2 and am.
prd-6 Linked to ad-7. Between inl (9%; 30%)
and pab-2 (2%).
Prf Linked to al-3 (3%), probably to the right.
pro-3 Linked to inl, pab-1 (0/74). Between
his-1 (4%) and pk (2%; 6%).
psp Between al-3 (10%; 22%) and his-6
(22%; 24%).
pvl(PCNA) Linked to NOR.
pyr-6 Between asn (6%) and un-9 (2%).
rca-1 Linked to cyh-2 (0T/16 asci).
rDNA Between Tip-VL and mus-18 (0T/17
asci; 20kb), dgr-1. In the NOR..
rec-1 Between ro-4 (7%) and asn (5%).
rec-2 Between sp (1%; 6%) and am (2%).
ro-4 Linked to pab-2 (0/407). Between ad-7
(4%), T(EB4)R and inv (5%).
rol-3 Between ilv-1 (2%) and cot-4 (5%).
sat At Tip-VL, distal to NOR.
scon-1 Probably linked to his-6.
scot Between al-3 (7%) and his-6 (11%).
sdv-6 Linked to crp-4 (0T/18 asci). Right of
inl (9T/18 asci).
sen Between his-1 (4%) and al-3 (13%).
ser-2 Between met-3 (4%) and
T(EB4)L, cot-2 (5%, 8%).
sh Between ilv (4%; 7%) and md (3%).
smco-6 Linked to asn (6%) near pyr-6.
smco-7 Right of ilv-1 (2%). Linked to rol-3
(0/154), sp (12%).
sod-2 Linked to cyh-2 (0T/18 asci.
sor-5 Linked to his-1.
sp Between leu-5 (3%; 9%) and ure-2 (8%),
am (1%; 8%; 450 kb).
spco-9 Linked to asn (6%); right of met-3
(18%).
spco-10 Between ilv-1 (24 and inl (5%).
spe-1 Between am (1%) and his-1 (1%) .
spe-2 Left of am (6%), spe-1.
spe-3 Right of inl (4%).
ssu-6 Linked to his-1 (4%).
su(het-d) Near lys-1.
su(ile-1) Right of met-3 (4%).
su([mi-1])-f Left of inl (10%).
T(UK1-35) Breakpoint in NOR.
T(UK2-32) Breakpoint in NOR.
T(UK3-41)L Between his-1
and al-3.
T(UK3-41)R Between pyr-6 and oak.
T(EB4)L Between pk, ser-1 and cot-2,
wa.
T(EB4)R Between ad-7 and
ro-4, pab-2.
T(UK4-22) Breakpoint in NOR.
T(DBL9) Between oak and Tip-R. Left of
his-6 (0/48).
T(UK14-1) Breakpoint in NOR.
T(AR30) Between Tip-VL and caf-1.
T(AR33) Between caf-1 and lys-1.
T(R55) Breakpoint in NOR.
T(MB67)L Between caf-1
and lys-1.
T(MB67)R Between cyt-9 and
Cen-V.
T(NM149) Linked to his-6 (0/499).
T(NM169d) Linked to NOR at Tip-VL.
T(ALS176) Breakpoint in NOR.
T(ALS182) Breakpoint in NOR.
T(AR190) Breakpoint in NOR..
T(OY321) Breakpoint in NOR.
Tel-VR Linked to his-6 (30kb).
Tip-VL Marked by sat, NOR, rearrangements.
Left of mus-18, dgr-1.
Tip-VR Marked by T(NM149). Right of his-6.
tom40 Linked to cyh-2 (0T/15 asci).
tom70 Right of inl (6 or 7T/17 asci).
trp-5 Between inl, pab-1 and
met-3 (4%).
ts Linked to inl (4%).
uc-4 Between inl (12%) and his-6 (11%).
udk Left of uc-4 (29%).
un-9 Between pyr-6 (3%) and oak (7%),
his-6 (5%; 7%).
un-11 Left of his-1 (<1%).
un-19 Linked to al-3 (9%), un-11 (14%).
ure-1 Between ace-5 (<1%) and his-1
(<1%).
ure-2 Between sp (8%) and am (2%),
ure-1 (3%).
usg-1 Linked to am (3.5kb).
val Linked to ilv-2 (0/135).
vma-1 Between leu-5 (1/18 asci), sp
(8%) and am (2%; ~40kb), al-3 (1/18 asci).
vma-3 Between his-1 and al-3. Linked to
leu-5 (0T17 asci), vma-1 (~1100kb), cyh-2.
wa Between pk,
T(EB4)L and ad-7 (4%).
Return to the Neurospora crassa genetic map
page
Return to the FGN 47 page
Return to the main FGN page
Return to the FGSC main page
Last modified 8/3/00 KMC