| 5YEG | |
| Yeast extract | 5 g |
| D-glucose | 10 g |

Susan G. W. Kaminskyj. Dept. Biology, Univ. Saskatchewan, 112 Science Place, Saskatoon, Saskatchewan, CANADA S7N 5E2.
This is a compendium of protocols for using Aspergillus nidulans in genetic, molecular, and cell biological investigations, originally written for members of my research group. It also summarizes our common growth media and nutritional supplements, many of which originally appeared elsewhere but now are difficult to locate. All solutions, tools, etc. are assumed to be sterile. All water should be sterile distilled or equivalent. Temperatures are in °C. Strains are available from the Fungal Genetics Stock Center (http://www.fgsc.net/).______________________________________________________________________________
Nomenclature Aspergillus nidulans is a deuteromycete, that is, a member of a polyphyletic group of fungi whose sexual stages are rare or unknown. If known, the name of the sexual stage (the teleomorph) has taxonomic precedence over the asexual stage (the anamorph), so this species is most properly known as Emericella nidulellus. Nevertheless, A. nidulans remains in common use in science and industry as an informal, inclusive name, which will be used here for both the anamorph and teleomorph. Growth and storage of Aspergillus nidulans conidia A. nidulans conidia germinate in defined (MM) or complex (e.g. CM and 5YEG) media, plus appropriate nutritional supplements for auxotrophic strains. See recipes below for details. Wildtype germination is best between 28° and 37°, range 2° to 48°. Some strains are temperature (heat) or cold sensitive. Germination is faster on complex than minimal media, and is favored by high inoculum levels. This is noteworthy when preparing protoplasts for transformation: a billion conidia germinate in 4-7 hours (strain and condition dependent) in 50 ml of liquid medium, but can take twice as long in 250 ml of medium. Monitoring germination shows that the conidia clump 30-60 minutes before producing a germ tube. Similarly on agar plates, clumps of conidia germinate and grow faster than isolated ones. A. nidulans ascospores typically require 2-3 days to germinate in the same conditions as conidia, and do not require special induction. Ascospores remain viable for several years in water at 4°C. Ascospore protocols are described in the section on mating. Harvesting A. nidulans conidia is best done in still air, not in a flow hood or a fume hood, because the spores are readily dispersed by air currents. Equally important, opening or closing a Petri plate creates turbulence within and should be done carefully and as seldom as possible. We leave plates open for extended periods while replica plating individual colonies with toothpicks. If contamination from lab air is a problem, air cleaners that have HEPA filters will make a noticeable improvement over a few weeks, and are widely available. A. nidulans conidia are harvested in water (by rubbing the surface of a conidiating culture with a sterile bent glass rod to dislodge the spores) and can be stored for weeks to months at 4°C without a serious loss of viability. However, freshly harvested spores germinate more reproducibly. For long-term storage, spores can be stored in 50% glycerol at –20°C, or in 15-20% glycerol at –80°C. Glycerol is more easily pipetted as an 80% solution in water. 800µl of aqueous conidial suspension (as made for 4°C storage) added to 200µl 80% glycerol is used for a –80°C stock; 400 µl suspension added to 600 µl 80% glycerol is used for a –20°C stock. Vortex before freezing. For mutant collections, small pieces of conidiating cultures can be excised and placed in 20% glycerol, vortexed, and frozen as –80°C stocks. A. nidulans conidia can be adsorbed onto sterile dry silica gel by making a spore suspension in sterile 4% non-fat milk, adding about 1 ml of spore suspension to 10 ml silica, shaking vigorously, and cooling in an ice bath (this process is exothermic). The silica should appear dry within a few hours. Conidia will remain viable for several years at room temperature or longer at 4°C. Cultures on agar plates that dry slowly at 4°C also have viable conidia. Sealing plates with Parafilm before they are completely dry can result in contamination. For reviving stored cultures, chips of –80°C medium, or vortexed –20°C or 4°C stocks should be streaked for single colonies on appropriate media. Silica stocks are sprinkled onto agar media.| Complete Medium (CM) | |
| D-glucose | 10 g |
| Peptone | 2 g |
| Yeast extract | 1 g |
| Casamino acids | 1 g |
| 20X nitrate salts (see below) | 50 ml |
| Trace elements (see below) | 1 ml |
| Vitamin solution (see below) | 1 ml |
Add nutritional supplements if necessary. Adjust pH to 6.5 with 1 M NaOH. Make to 1 liter with distilled water. For solid medium add 15 g agar. Autoclave.
| Minimal Medium (MM) | |
| D-glucose | 10 g |
| 20X nitrate salts (see below) | 50 ml | Trace elements (see below) | 1 ml |
| 1% thiamine | 1 ml |
Add nutritional supplements if necessary. Adjust pH to 6.5 with 1M NaOH. Make to 1 liter with distilled water. For solid medium add 18 g agar. Autoclave.
|
 |
||||||
Add nutritional supplements if necessary. Make to 1 liter with distilled water. For solid medium add 18 g agar. Autoclave. Note that green-spored paba mutants (FGSC A28 is shown) are yellow on 5YEG.
| 20 X Nitrate salts | |
| NaNO3 | 120 g |
| KCl | 10.4 g |
| MgSO4·7H2O | 10.4 g (5.2 g if anhydrous) |
| KH2PO4 | 30.4 g |
Add distilled water to 1 liter. Autoclave. Store at 4oC.
| 1000 X Trace elements | |
| distilled water | 60 ml |
| ZnSO4·7 H2O | 2.2 g |
| H3BO3 | 1.1 g |
| MnCl2·4 H2O | 0.5 g |
| FeSO4·7 H2O | 0.5 g |
| CoCl2·6 H2O | 0.17 g |
| CuSO4·5 H2O | 0.16 g |
| Na2MoO4·2 H2O | 0.15 g |
| Na4EDTA | 5 g |
Vitamin Solution
Add 100 mg each of biotin, pyridoxin, thiamine, riboflavin (light sensitive), PABA (p-aminobenzoic acid), nicotinic acid to 100 ml water. Add 2 drops chloroform as preservative. Store in a dark bottle at 4°C.
Nutritional supplements for our common auxotrophies
Autoclave and store at 4°C. These and others were originally quantified by Etta Käfer. Amino acids should be the L-isomer. The salt form dissolves more readily.
| Stock g/100 ml | Volume Stock per 200 ml | Volume Stock per liter | |
| adenine | 0.86 g | 2 ml | 10 ml |
| arginine | 4.2 g | 1 ml | 5 ml |
| biotin (in 70% ethanol, –20C) | 0.05 g | 10 µl | 50 µl |
| choline | 4.1 g | 100 µl | 500 µl |
| cysteine | 0.4 g | 1 ml | 5 ml |
| isoleucine | 0.6 g | 2 ml | 10 ml |
| leucine | 0.6 g | 1 ml | 5 ml |
| methionine | 0.5 g | 1 ml | 5 ml |
| nicotinic acid | 0.5 g | 100 µl | 500 µl |
| ornithine | 3.4 g | 1 ml | 5 ml |
| para-amino benzoic acid (PABA) | 1 g | 200 µl | 1 ml |
| phenylalanine | 1 g | 1 ml | 5 ml |
| proline | 1 g | 1 ml | 5 ml |
| pyridoxin (pyro) | 0.1 g | 10 µl | 50 µl |
| thiamine | 0.25g | 200 µl | 1 ml |
| urea | 6 g | 1 ml | 5 ml |
Atypical supplements
Pyrimidines (uridine + uracil): uridine 1.2 g solid /liter + uracil 1.12 g solid/liter. Pyrimidine stock: 5.6 g uracil + 6.1 g uridine/100 ml. Use 20 ml/l. Looks like lumpy milk which dissolves when diluted or warmed. Shake well, then measure with a graduated cylinder. Tryptophan: 1g solid/100 ml. Add to warm medium after autoclaving. Ampicillin stock, 1000x From sodium salt: 0.5 g ampicillin in 10 ml 70% ethanol. Stir until dissolved and store at –20°C. From trihydrate: 0.5 g ampicillin in 3 ml 1 M NaOH. Stir until dissolved. Add 7 ml 100% ethanol and store at -20°C. The solution will be yellow but still effective. Mating and analysis of A. nidulans meiotic progeny Mate strains by growing mixtures of spores in fully supplemented liquid medium for 2 days at 28°C. Strains must have complementary auxotrophies to form hyphal anastomoses efficiently for heterokaryon (and later for cleistothecium) formation. However, prototrophs can mate, and strains can self-mate. Where possible, use different spore colors for the two parent strains as this will make it easier to check whether both parents are growing, and later will be useful for identifying outcrossed cleistothecia.
 It is most convenient to use a 24 well tissue culture dish for matings, but it is
also possible to use sterile Eppendorf tubes. For tissue culture dishes, use about 2 ml of fully supplemented liquid
medium per well. Typically we use two wells for each parental combination, adding 1 µl of each conidial stock per well.
Ideally, similar numbers of fresh spores should be used for each parent. If one strain does not grow adequately, it can be
reinoculated into the original well, and incubated another day. Alternatively, additional wells can be prepared with
increasing numbers of the lagging strain.
It is most convenient to use a 24 well tissue culture dish for matings, but it is
also possible to use sterile Eppendorf tubes. For tissue culture dishes, use about 2 ml of fully supplemented liquid
medium per well. Typically we use two wells for each parental combination, adding 1 µl of each conidial stock per well.
Ideally, similar numbers of fresh spores should be used for each parent. If one strain does not grow adequately, it can be
reinoculated into the original well, and incubated another day. Alternatively, additional wells can be prepared with
increasing numbers of the lagging strain.
For Eppendorf cultures, co-inoculate the parental strains in 400 µl liquid medium, close tightly and place loose in a small Erlenmeyer flask in a shaking incubator for 24 h at 28°. Remove tubes to an Eppendorf rack, poke a hole in the lid using a hot needle, and incubate as a stationary culture for another day.
When mating a cold-sensitive (cs) strain with wildtype, we have had good results from allowing the cs strain to grow at 37° for 1-2 days before adding wildtype spores and shifting to 32° for another day. Similarly, a poorly conidiating strain can be mated by growing hyphal explants with spores from the other parent. Allow the parental mycelium to grow just until it has pigmented spores. Transfer the hyphal mat using forceps to an MM plate (if the parent strains share any auxotrophies, these must be supplemented) and incubate at 28°C for 1-2 d. Removing the mat intact is easier if the forceps are run around the edge of the well to loosen the hyphae. Cleistothecia form most abundantly where the mat is in contact with the MM agar; we cut the edges so it will lay flat. Some people dip their hyphal mats in water, and then drain/blot dry to remove nutrients, but we have not found this to be necessary. We routinely put two or more (for poor-mating combinations) hyphal mats per MM plate. After 2 days, seal the plate with Parafilm (fold a 3 cm wide piece of Parafilm in half and seal with the double layer) to prevent the medium from drying out, and store in the dark for 2-3 weeks for the cleistothecia to mature. Cleistothecia become noticeably more fragile after 6-8 months, but the spores remain viable and can still be used for isolating double mutants. Outcrossed cleistothecia are found more reliably on the hyphal mat rather than in the surrounding heterokaryon. They are large and found near conidia with each of the parental colors. Sometimes it is possible to see both parental spore colors on a single conidial head of a heterokaryotic strain. There is considerable variation in size and abundance of cleistothecia for any one cross, and variation in average cleistothecium size for different strain combinations. Typically, mature A. nidulans cleistothecia have a shiny black peridium covered with a layer of light yellow Hülle cells, which may be viable, and plus parental conidia. These are removed by rolling the cleistothecium on a plate of 3% agar, a process made more efficient by adding 1% diatomaceous earth as a mild abrasive. Once clean, the cleistothecium is transferred to an Eppendorf tube containing 200µl water, crushed, and vortexed to release the ascospores. These are crimson, and make a red spot on the bottom of the tube upon settling.
Before analysis, the ascospores from individual cleistothecia should be tested for evidence of outcrossing. A loopful of ascospore suspension is spread over a small area of a fully supplemented plate, and incubated until the colonies have pigmented conidia. These should have at least both parental colours, plus recombinants if possible from the cross. See discussion of conidial color. Here are test spreads from four cleistothecia of a green:white mating. Although all appear to be outcrossed, given the choice we would choose not to analyze the cleistothecium whose spread in the 3 o’clock position since the color does not appear to be segregating 1:1. This is a convenient stage to test complementation vs allelism. Genes from different complementation groups will produce wildtype progeny with all possible colors under selective conditions.

Outcrossed cleistothecia are routinely analyzed by random ascospore analysis; some labs use octad dissection, analogous to tetrad dissection in Saccharomyces. For random ascospore analysis, we make a 2:200 dilution of the ascospore stock, and spread 30µl, 60µl and 90µl of this on appropriately supplemented plates, as shown. This is a convenient stage at which to select against unwanted nutritional markers. At least one of the spore concentrations should be appropriate for picking isolated colonies. Spreading can be improved by adding 0.01% Triton X-100; if so, the diluted spores should be spread immediately as even overnight storage compromises viability. These plates were spread without using Triton X-100.
Even with low spore concentrations and Triton X-100, some isolated colonies can grow from two nearby ascospores, particularly if the asci weren’t disrupted thoroughly. These colonies are easy to identify if they have two colors, but since neighboring spores might produce the same color conidia, even single-colored colonies should be picked near an edge. Random ascospore analysis
To haploidize a putative diploid strain, spores are grown on a benomyl gradient to destabilize mitotic nuclei. Spread spores of the putative diploid on a plate, and then apply benomyl suspension directly to the agar surface. Explants from the growth margin are transferred to fully supplemented benomyl-free plates. The left figure shows the effect of 1-2 µl drops of 10mg/ml benomyl suspension (white spots in the center) in 95% ethanol after 2 and 3 days at 37°. Sectored colonies produced in this way are shown on the right.
Mitotic mapping - assigning genes to chromosomes
Make diploids between a strain with your favorite gene (yfg) and FGSC A104 or another strain with a marker on each chromosome (see http://www.fgsc.net). The A104 genotype is yA2 (yellow spores), adE20 (adenine requiring), both Chr I; AcrA1 (resistance to acriflavin) Chr II; phenA2 (phenylalanine requiring) Chr III; pyroA4 (pyridoxin requiring) Chr IV; lysB5 (lysine requiring) Chr V; sB3 (sulfate non-utilization, methionine requiring) Chr VI; nicB8 (nicotinamide requiring) Chr VII; coA1 (compact colony) Chr VIII. Depending on the yfg phenotype, coA1 can be difficult to score; for example, yfg strains containing pyrG89 are compact even on fully supplemented media. However, chaA1 (chartreuse spores) is also on Chr VIII, and can be scored if the yfg strain is chartreuse.
Factors when choosing the yfg parent for mitotic mapping with A104:
a) Yellow strains are not appropriate. b) Green strains are not optimal, since most sectors will be green, and will vary only in subtleties of shade or fluffiness. Neighboring sectors with the same spore color can sometimes be identified through the bottom of the plate. Ideally at least 100 independent haploid sectors should be picked for this analysis so obvious color differences are an advantage. c) The yfg strain should not have an auxotrophy found in A104, which will prevent scoring results for that chromosome d) The best choice is a chartreuse yfg strain, since ChrVIII can be scored by spore colour, and there will be many variations in sector color. e) Second best is a white yfg strain, since there will be at least three sector colors, but Chr VIII must be scored by compact colony morphology. Pick all the sectors onto a fully supplemented CM master plate (28°C) and an MM plate to test for diploid sectors. The fully supplemented medium must support all the A104 auxotrophies plus those in the yfg parent. Include a colony from each parent on each plate, which makes scoring +/– growth easier. Replica pick from the master plate to selective media and to a fully supplemented plate at 42°C if yfg is ts-. For scoring the nutritional markers, the selective plates will be MM supplemented for all yfg strain auxotrophies plus all but one A104 auxotrophies. To score for AcrA1, use CM plus all supplements, plus acriflavin (0.5g/100ml stock, 5ml stock per liter medium; 2.5 mg acriflavin solid per liter), added after autoclaving. Only AcrA1 strains grow on this medium. Interpreting the data: Assume for this discussion that yfg is ts-. For 7 of 8 chromosome markers, each marker should be associated with similar numbers of ts+ and ts- sectors. This pattern holds for the chromosomes that do not have yfg. For the remaining chromosome, a sector with the A104 marker should never be ts- (for exceptions, see below). In other words, yfg will segregate "in repulsion" from this chromosomal marker. One exception to this is when yfg is on Chr VIII and the yfg parent has chartreuse spores. In this case, yfg will segregate "in coupling" with chaA1. So, if your strain of yfg is temperature sensitive, ts- sectors are evidence against yfg being associated with a specific chromosomal marker. However, ts+ sectors are neutral. Mitotic crossing over between sister chromatids can skew the data, even assuming that all the sectors are properly scored for all markers, but should be relatively rare. Common spore colors in Aspergillus nidulans Wildtype alleles are written as +, and "::" means mated to. Typically wildtype alleles are implied, but here they are written explicitly. Wildtype conidia are dark green; commonly used color mutants are white, yellow, chartreuse, but there are others such as fawn. Spore pigmentation is controlled by wA, wildtype allele wA+. Strains with the mutant allele wA2 have white spores because the defect prevents pigment formation.Wildtype dark green pigment requires yA+. Strains with wA+ can make pigment, but strains that are wA+, yA2 have yellow conidia. Strains that are wA2, yA2 are white because wA2 is epistatic to yA2. If a white strain and a yellow strain are mated, they can produce green progeny (see the ascospore spread picture, from a white / yellow strain mating) since these are independent loci.
Chartreuse spores are caused by a defect in a modifier gene, chaA1 that dilutes the color formed otherwise. So, wA+, chaA1 spores are bright green rather than dark; as before, wA2, chaA1 spores are white. The spores of wA+, yA2, chaA1 strains are light yellow. A wA2 (white) strain can have a color allele such as chaA1 in its genetic background. If such a strain is mated to a wA+ (green) strain, gene segregation patterns will reveal if it has chaA. A green :: white = wA+, chaA+ :: wA, chaA, mating produces four progeny genotypes: wA+, chaA+ (green); wA+, chaA1 (chartreuse); wA2, chaA+ (white); wA2, chaA1 (white). The phenotype ratio is green : chartreuse : white = 1 : 1 : 2.
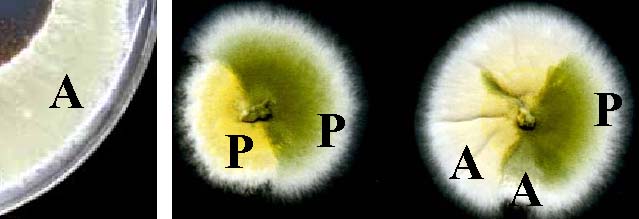
Some auxotrophies can affect spore color. Here, arginine auxotrophs (A) and prototrophs (P) are growing on fully supplemented medium.
Light microscopy Inoculate conidia (5 ml of 5 x 104 spores/ml) into liquid medium with nutritional supplements and 100 µg/ml ampicillin, in 60 x 15 mm Petri dishes containing one coverslip. Coverslips can be used straight from the box if handled with forceps. Incubate as required. Pour off the medium; the coverslip and attached germlings will be retained by surface tension. If the germlings grow too long, they can shear off since their attachment is at the conidium only.Fix germlings by flooding with 3.7% formaldehyde in 50 mM potassium phosphate buffer, pH 7.0, containing 0.2% Triton X-100. A 5x stock of fixative is made by mixing 50 ml 37% formaldehyde solution, 50 ml 500mM potassium phosphate buffer pH7, 2 ml Triton X-100. Rinse fixed specimens with distilled water (optional).
Stain nuclei with Hoechst 33258 (Sigma bis-benzmide B2883) and cell walls with Calcofluor. Both are excited by UV and have blue-white fluorescence. Both stain stocks are 1 mg/ml in water. Calcofluor dissolves completely under alkaline conditions (~25 µl 0.1M NaOH in 10 ml), but this does not improve staining. The optimum amount of stain varies with strain and growth conditions. Our typical working solution is 16 µl Hoechst 33258 and 8µl Calcofluor in 50 ml of water. Store Calcofluor stock at 4°C in the dark; store Hoechst 33258 stock at –20°C. Make the diluted staining solution shortly before use and stain for at least 1 min. Rinse at least twice with distilled water after staining to reduce background. Mount in antifade solution: 50mM Na-phosphate buffer, pH 8.8, containing 50% glycerol and 0.1% n-propyl gallate. This solution is almost colorless when freshly made, and should be replaced when it turns yellow. Seal slides with nail polish and store at 4°C in the dark. Fluorescence will be stable ~ 2 months, and often is better the day after the slides are made. Hoechst 33258 appears to be trapped non-specifically in cell walls, which is useful for dual localization in thick-walled cells where the Calcofluor signal can obscure nuclei. Immunofluorescence protocol for Aspergillus Grow spores as described for light microscopy. Fix 30-60 min at room temperature in formaldehyde/MBS/P+BS (recipe below). Wash 3 x 5 min in P+BS. Partially degrade the hyphal walls in enzyme solution (recipe below), 200ul per coverslip, 30-90 min, by inverting the coverslip over a droplet on Parafilm. The time will vary with the strain, antibody, and growth conditions. Wash 3 x 5 min in P+BS. Permeabilize membranes in 0.01% Triton X-100 (most antibodies) or –20°C methanol (antiactin) for 20 min. Solvents can interfere with binding in many antibody/epitope combinations. Block nonspecific binding with freshly made 5% nonfat milk in P+BS for 20 min. Incubate in 20-100µl primary antibody dissolved in 0.5% nonfat milk (1:9 blocking solution: P+BS) for 30-90 min, germling-side up on a Parafilm sheet. Cover with a Petri dish/damp tissue in the lid, to prevent evaporation. To determine appropriate antibody concentrations, first use a range of primary antibody concentrations (1:10, 100, 1000...) with a constant secondary antibody conc (1:10). Once the optimum primary antibody concentration is determined, repeat with varying secondary antibody concentrations (1:10, 50, 100, 200). Some monoclonal antibodies give a good signal at 1:100,000 but others are used undiluted! Using too high a primary antibody concentration gives unacceptably high background. Wash 3 x 5 min in P+BS. This is the minimum time for washes, which are important for removing trapped primary antibody that otherwise contributes to non-specific staining. Incubate in 100µl of species-matched fluorescently labeled secondary antibody dissolved in 0.5% nonfat milk in P+BS for 30-90 min. Cover with a Petri dish/damp tissue in the lid, to prevent evaporation. Incubate in the dark to minimize fluorochrome fading. Wash 3 x 5 min in P+BS. Counterstain with Calcofluor and/or Hoechst 33258 (optional). Mount in antifade, 5 µl per coverslip. Immunofluorescence preparations are best examined the day they are made. Fixative 3.7% formaldehyde in P+BS, containing 100uM MBS (stored in dessicator at 4°C, as 100 mM in DMSO; warm to room temperature before using). Fixative can be made from stock 37% formalin, but it is better to make it fresh from paraformaldehyde crystals. Dissolve the crystals at 2X final concentration in distilled water containing a drop 0.1N NaOH, with gentle heating, and then add 10x P+BS and water. MBS is m-maleimidobenzoyl N-hydroxysuccinimide ester (Sigma, Molecular Probes), a secondary crosslinking agent that improves preservation and stability of formaldehyde-fixed cells. Unlike glutaraldehyde, it does not cause autofluorescence of fixed specimens. P+BS PIPES + buffered saline, pH6.8, 10x stock, 100ml. Store at 4°| PIPES (50 mM) | 15.12 g |
| EGTA (2mM) | 0.760 g |
| MgSO4 (2mM) | 0.241 g |
| NaCl (137 mM) | 8 g |
| KCl (268µM) | 0.2 g |
Enzyme solution: store in aliquots at –80°
250 mg Driselase (Interspex http://www.wenet.net/~sharma/interspex/)
250 mg Glucanex (Interspex)
250 mg Beta-glucuronidase (Sigma)
250 mg BSA
250 mg non fat milk
1 mM AEBSF (see below)
5 ml P+BS
AEBSF is a protease inhibitor sold by Calbiochem and others. Unlike PMSF, it is stable indefinitely in aqueous solution; PMSF is effective for 5 min in aqueous soln. Make a stock of 500mM AEBSF in aqueous solution, and store at room temperature or 4°. Both inhibitors are effective against serine-threonine proteases at 1 mM.