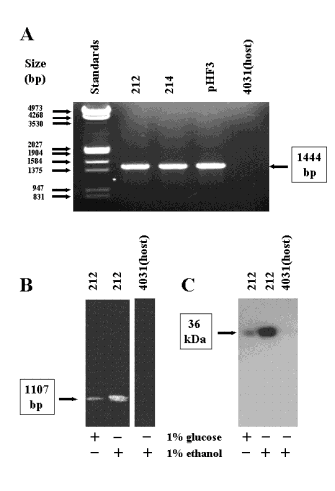
Figure 1. Transformation and expression of the A. nidulans bimG gene in N. crassa
Expression of the Aspergillus bimG gene in Neurospora crassa
Balázs Csóka1, Tamás Zeke1, Endre Kókai1, John Doonan2, Helen Fox2, Zsigmond Fehér3 and Viktor Dombrádi1
1Department of Medical Chemistry, Medical and Health Science Center, University of Debrecen, Debrecen, Bem tér 18/B, H-4026, Hungary, 2John Innes Center, Colney Lane, Norwich NR4 7UH, UK, 3Department of Human Genetics, Medical and Health Science Center, University of Debrecen, Debrecen, Nagyerdei krt. 98, H-4012, Hungary
Fungal Genet. Newsl. 49:13-14
In A. nidulans the bimG gene codes for the catalytic subunit of protein phosphatase 1. The wild type bimG gene was transformed into N. crassa and expressed under the direction of the alcA promoter. The heterologous bimG mRNA and protein were detected in the transformants by RT-PCR and Western blotting, respectively. However, the transformation did not result in detectable changes in phenotype. This work demonstrates that the alcA promoter, a conditional gene expression system widely used in both Aspergillus and higher plants, also functions in N. crassa.
BimG11 was described as a temperature-sensitive recessive mutation that causes the block of mitosis accompanied with the over-phosphorylation of nuclear proteins and distinct morphological changes in Aspergillus nidulans (Doonan and Morris, 1989 Cell 57: 987-996). In addition the mutant was defective in cell-wall synthesis at the restrictive temperature (Borgia, 1992 J. Bacteriol. 174: 384-389). Molecular cloning revealed that the bimG gene encoded a protein similar to the catalytic subunit of protein phosphatase 1 (PP1c). In agreement with the structural prediction, PP1 activity was reduced in bimG11 mutants at 40oC (Doonan et al., 1991 J. Biol. Chem. 266: 18889-18894). A single point mutation near the splice-site of intron 2 resulted in the translation of a truncated inactive phosphatase at higher temperature, which was supposed to exert a toxic effect (Hughes et al., 1996 EMBO J. 15: 4574-4583). The rabbit skeletal muscle PP1ca cDNA rescued the mutant phenotype, while a AtPP1bg coding for a PP1c isoenzyme of Arabidopsis thaliana affected partial complementation (Arundhati et al., 1995 Plant J. 7: 823-834). The cDNA and gene of PP1c (termed ppp-1) has been recently cloned from Neurospora crassa in our laboratory (Zeke et al. unpublished results). The predicted amino acid sequence of the PPP-1 protein is highly similar to the bimG gene product (96% identity). In order to study the possible application of the alcA promoter based conditional expression system in another filamentous fungus, we expressed the wild type bimG from A. nidulans in N. crassa.
We constructed a plasmid (termed pHF3) in which the wild type bimG gene was fused at the carboxy-terminus to an influenza haemaglutinin (HA) tag and was placed under the control of the A. nidulans alcA (alcohol dehydrogenase) promoter. The construct harbored the pyr-4 gene of N. crassa that was used as a selection marker. A second plasmid (pSRN1) encoding for the AlcR regulatory protein was also used to ensure the efficient gene expression from the alcA promoter. A pyr-4 strain of N. crassa (FGSC 4031) was co-transformed with the above two plasmids as described by Vollmer and Yanofsky (1986, Proc. Natl. Acad. Sci. USA, 83: 4869-4873). The transformants were grown at 27°C for 24 hours in liquid Vogel’s minimal medium supplemented either with 1% (v/v) ethanol (inducing the alcA promoter) or with 1% (w/v) glucose (repressing condition). Hyphae were harvested by filtration, frozen in liquid nitrogen and stored at -70°C until use.
The incorporation of the A. nidulans bimG gene into the N. crassa genome was demonstrated by polymerase chain reaction (PCR). The forward primer: CTTCGTTGGGGGTAAAACGCC hybridized to the 5’-noncoding region of the bimG gene, while the reverse primer: ATCTAGAGCGGCCGCACTGAG was specific for the HA tag. One microgram of genomic DNA prepared according to Sambrook et al. (1989, Molecular cloning, 2nd edition, Cold Spring Harbor Laboratories) was amplified with Accu Taq polymerase (Sigma-Aldrich) in 30 cycles according to the following temperature profile: 0.5 min at 94 oC, 1 min at 62 oC, and 1 min at 68 oC. The expected 1444 bp band was detected by ethidium bromide staining in two transformants after agarose gel electrophoresis in 1 % (w/v) agarose gels (Fig 1. A).
The expression of the heterologous bimG gene was analyzed by PCR after reverse transcription (RT-PCR). Reverse transcription was performed for 45 min at 48 oC with the enhanced avian RT-PCR kit of Sigma-Aldrich. The same primers and temperature profile were used as in the previous PCR experiment for the amplification of 0.3 micrograms of total RNA prepared according to Chomchinsky and Sacchi (1987 Anal. Biochem. 162: 156-159). The 1107 bp bimG specific cDNA was visualized with ethidium bromide (Fig. 1. B). Obviously, the control of expression was leaky in the No. 212 transformant, since the mRNA band was detectable even in the presence of glucose. The intensity of the band was elevated under ethanol induction conditions. The bimG mRNA was hardly detectable in the No. 214 transformant (not documented) and was absent from the host strain FGSC 4031 (Fig. 1. B).
Western blotting (Harlow and Lane 1988 Antibodies, Cold Spring Harbor Laboratories) was used to demonstrate the translation of the bimG mRNA. Fifty micrograms of protein extracted from the No. 212 transformant was separated by SDS PAGE in a 10% (w/v) gel and transferred onto a HybondTM ECLTM nitrocellulose membrane (Amersham Life Sciences). The blot was blocked, chalenged with an anti-HA monoclonal antibody (HA.11, BAbCO), and visualized by a peroxidase conjugated anti mouse IgG second antibody (Sigma) and ECL technique (Fig. 1.C). In accordance with the RT-PCR results a 36-kDa band of the A. nidulans BimG protein was visible in the presence of glucose, and the expression of the protein was more pronounced after the addition of ethanol. The host strain exhibited no cross-reactivity with the antibody. Our results indicate that the alcA based expression system operates in N. crassa and is moderately inducible by ethanol at both the mRNA and protein levels. The heterologous expression of the bimG gene also implies that splicing signals of the foreign gene were functional in N. crassa.
Protein phosphatase 1 (PP1) activity was assayed with 32P-labeled rabbit muscle phosphorylase a in the presence and absence of inhibitor-1, a specific inhibitor of PP1 (MacKintosh, 1993 in Protein Phosphorylation, pp. 197-230, Oxford University Press). The PP1 activity in the extract of the control FGSC 4031strain was estimated to be 1.21 + 0.23 mU/mg (n=14) in the presence of 1% (w/v) glucose, and 1.30 + 0.31 mU/mg (n=20) in the presence of 1% (v/v) ethanol. In the No.212 transformant the corresponding values were 1.25+ 0.16 mU/mg (n=19) in the presence of glucose and 1.25 + 0.19 mU/mg (n=13) under inducing conditions. Thus the expression of the heterologous phosphatase catalytic subunit had no significant effect on the basal PP1 activity of N. crassa. The lack of activity change can be explained either by the relatively low level of expression, or by the operation of a compensatory mechanism eliminating the effect of the surplus A. nidulans phosphatase. The transformant exhibited normal growing behavior and no visible changes of morphology were discerned by microscopic observation. In agreement with the latter findings ectopic expression of BIMG phosphatase in Aspergillus has no phenotypic consequences for growth or development and does not significantly alter the PP1 activity levels (Doonan, unpublished data).
Thanks are due to Dr. Oded Yarden (Hebrew University of Jerusalem, Rehovot, Israel) for the examination of the morphology of the N. crassa transformants. Our work was supported by the OTKA 33050 grant of the Hungarian Research Foundation.

Figure 1. Transformation and expression of the A. nidulans bimG gene in N. crassa
A. PCR detection of the bimG gene in No. 212 and 214 N. crassa transformants. The plasmid pHF3 was used as a positive control. B. RT-PCR analysis of the bimG mRNA in No. 212 N. crassa transformant. C. Western blot analysis of the BimG protein in the No.212 N. crassa transformant. The host strain FGSC 4031 served as a negative control in all experiments. Representative results of three independent experiments are shown.
Return to the FGN 49 Table of Contents
Return to the Main FGN page
Return to the FGSC Main page