
A quick method to isolate pure DNA from asexual spores of Coprinus cinereus for screening approaches
Piers J. Walser1, U. Kues1,2 and M. Aebi1
1 Institute for Microbiology, ETH Zurich, Schmelzbergstr. 7, CH-8092 Zurich, Switzerland
2 Institute for Forest Botany, Georg-August-University Gottingen, Busgenweg 2, D-37077 Gottingen, Germany
Fungal Genet. Newsl. 49:15-16
In this study, we present a glass bead-phenol method to isolate genomic DNA from oidia of the basidiomycete Coprinus cinereus. The DNA can be used in Southern blot analysis with digoxigenin-labelled DNA probes without the background problems encountered with DNA isolated from fungal mycelium. Furthermore, DNA isolated from oidia can be applied in PCR. This is especially useful when searching for specific DNA sequences or recombination events in a mixture of different strains.
Three methods have been published to isolate genomic DNA from vegetative mycelium of the basidiomycete Coprinus cinereus. A longer maxi-prep has been described by Wu et al. (1983 Curr. Genet. 7: 385-392) and Mutasa et al. (1990 Curr. Genet. 18:223-229). High-molecular weight DNA can be purified by CsCl-gradient centrifugation or, alternatively, it might be harvested from the solution with a glass hook. DNA obtained by the maxi-prep method is excellent for construction of genomic libraries (Bottoli et al. J. Microbiol. Meth. 35:129-141). The mini-prep method from Zolan and Pukkila (1986 Mol. Cell. Biol. 6:195-200) uses small amounts of lyophilised mycelium in an Eppendorf tube. In comparison, it is fast but gives yields of only 8-10 ?g DNA. In DNA Southern blot screening approaches, this amount of DNA is usually enough to obtain the required results (Granado et al. 1997 Mol. Gen. Genet. 256:28-36). However, in our lab we needed to use radioactively labelled probes in hybridisation, and impurities of unknown character caused a strong background smudging specific signals when probes were labelled with the non-radioactive DIG (digoxigenin) method (Roche Diagnostics Corp., No. 1277 065). In an effort to overcome the need for radioactivity, we developed an isolation procedure using the unicellular aerial spores (oidia) of Coprinus, a DNA isolation approach commonly used for single yeast cells (Hoffman and Winston 1987, Gene 57:267-72).
In short, the method is as follows: oidia from a single mycelial culture (~109 spores) were harvested by scraping the aerial mycelium with about 10 ml sterile water and, in the first experiments, the suspension was filtered through sterile glasswool to separate mycelial debris from the spores (this step could be omitted without any change in DNA yield). The spores were pelleted in a 50 ml Falcon tube, resuspended in 1.0 ml H2O and transferred into a microcentrifuge tube. Then 200 ml of extraction buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl pH 8.0, 300 mg/ml RNase A) and 100-200 ml of glass beads (425-600 mm diameter) were added and the tube either vortexed for 2 minutes or reciprocated for 20 seconds (speed 4) on a FastPrep (Bio101, FP120) machine to disrupt cells. Following disruption, tubes were briefly incubated on ice (up to 5 minutes) and finally 200 ml phenol:chloroform:isoamyl alcohol (25:24:1) were added and the tube carefully shaken until fully emulsified. The two phases were separated by centrifugation at 15,000 g for 5 minutes in a benchtop centrifuge. The aqueous phase (typically 250 ml) was ethanol precipitated by addition of 1/10 volume 3.0 M NaOAc pH 5.2 and 2 volumes of ethanol followed by incubation at -20? C for up to 20 minutes. The DNA was pelleted by centrifugation at 15,000 g for 10 minutes and the pellet washed once with 70% ethanol. The resulting pellet was briefly air-dried and dissolved in 50 ml H2O. Typically, 150 mg DNA was obtained in this procedure which is significantly more than the amount obtained by mycelial techniques. This increased yield was expected, because all asexual spores are nucleated with relatively little cytoplasm compared to mycelium. In contrast to the DNA obtained by the method of Zolan and Pukkila (1986), DNA isolated from oidia gave clear and clean signals with DIG labelled probes (Fig. 1).
Freedman and Pukkila (1993 Fungal Genet. Newsl. 10:36-37) published a PCR protocol for rapid screening approaches employing Zolan and Pukkila?s DNA mini-prep methods on mycelium grown directly in Eppendorf tubes. In our hands, we had problems with reliable growth of colonies in Eppendorf tubes, suggesting that the method is not as suitable for a high through-put approach, as needed for example when trying to isolate rare gene knock-outs by transformation. Albeit spores need to be harvested, DNA from oidia can be very useful in a PCR screening approach. Since merely a few template molecules are required in a DNA sample for identification by PCR, it is possible to grow a bunch of strains to small colonies on the same plate, harvest all oidia as a mixture from the plate, and isolate and analyse the DNA from the mixture of spores by PCR. In our test approaches, it was possible to detect the presence of a specific clone 30-fold diluted with wild-type spores (see. Fig. 2), i.e. it should be possible to pool individual transformants to an empirically derived degree (in this case = 30 transformants). By appointing a clever pooling strategy when growing the colonies, it will be very fast to identify a specific clone by a few rounds of PCR with oidia DNA.
Acknowledgements
We thank Jose Granado for his intense efforts to make DNA hybridisation with DIG-labelled probes and genomic Coprinus DNA work. We are grateful to the ETH Zurich, the Swiss National Foundation (grant 31-59157.99) and the DBU (Deutsche Bundesstiftung Umwelt) for financial support.
 |
| Figure 1: Southern blot analysis of gfp transformants of Coprinus strain AmutBmut (Swamy et al. 1984 J. Gen. Microbiol. 130: 3219-3224) hybridized with a gfp specific digoxigenin-labelled probe. Genomic DNA was digested with HindIII. Lane 1: gfp containing plasmid, lane 2:wild-type, lanes 3 -18: gfp transformants (lane 12 was not digested and the signal is outside of the sector of the blot shown) |
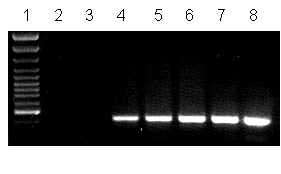 |
| Figure 2: Qualitative identification of a specific clone amongst others in PCR based screening (simulation of oidial dilution by pooling transformants, amount of template DNA not standardized). The primers used predict a gfp specific amplification product of 432 bp. Lane 1: 100 bp DNA ladder (Fermentas), lane 2: wild-type AmutBmut mycelium, lane 3: wild-type oidia, lane 4: oidia from gfp-transformant diluted 1:3 with wild-type oidia, lane 5: dilution 1:12, lane 6: dilution 1:30, lane 7: oidia from gfp-transformant, lane 8: gfp-containing plasmid used in transformation. |
Return to the FGN 49 Table of Contents
Return to the Main FGN page
Return to the FGSC Main page
