Haag, Jeremy R.1, Lee, Dong, W.2, and Aramayo, Rodolfo2,3. 1Department of Biology, Washington University, Campus Box 1137, 1 Brookings Drive, St. Louis, MO 63130. 2Department of Biology, Texas A&M University, Room 415, Building BSBW, College Station, TX 77843-3258
Fungal Genet. Newsl 50:6-8
We report the construction of a Destination Vector, called pJHAM007, for the targeted integration
of DNA sequences at the histidine-3 (his-3) locus of Neurospora crassa. pJHAM007 has all the necessary
features required to perform a simple, rapid and efficient GATEWAY™ recombinational cloning with an
Entry Clone to yield a his-3-gene replacement Destination Vector.
Gene replacement is a powerful tool to construct isogenic strains containing different DNA sequences integrated at the same chromosomal position. The most popular locus used for gene targeting in Neurospora crassa is of the metabolic gene histidine-3 (his-3). Several generations of plasmids for integration at this chromosomal position have been constructed (Sachs and Ebbole 1990 Fungal Genet. Newsl. 37: 35-36, Ebbole 1990 Fungal Genet. Newsl. 37: 15, Margolin, et al. 1997 Fungal Genet. Newsl. 44: 34-36, Aramayo and Metzenberg 1996 Fungal Genet. Newsl. 43: 9-13). Recently, we described the construction of a new set of N. crassa strains and plasmids that represent a significant improvement over previous systems because they allow the investigator to screen in one simple step for homokaryotic transformants containing the insertion of a test sequence among a population of primary histidine-independent transformants (Lee, et al. 2003 Curr. Genet. DOI 10.1007/s00294-002-0366-z). These new tools have significantly reduced the time it takes to construct new N. crassa strains. To expedite this system even further, we have designed and constructed a new plasmid, pJHAM007, that can be used for the high-throughput cloning of DNA inserts, to generate his-3-gene replacement plasmids for different types of large- or small-scale genome analysis.
Plasmid pJHAM007 is based on the GATEWAY™ system (Walhout, et al. 2000 Method. Enzymol. 328: 575-592). GATEWAY™ is a novel universal system for cloning and subcloning DNA sequences that uses phage lambda (λ)-based site-specific recombination (Landy 1989 Annu. Rev. Biochem. 58: 913-949). This Recombinational Cloning (RC) consists on two reactions: (1) The LR Reaction (attL X attR → attB + attP), mediated by the Integrase (Int), Integration Host Factor (IHF) and excisionase (Xis); and (2) the BP Reaction (attB X attP → attL + attR), mediated by the Int and IHF proteins. By providing different combinations of the recombination proteins and sites, the direction of the reaction can be easily controlled (Walhout, et al. 2000 Method. Enzymol. 328: 575-592, Hartley, et al. 2000 Genome Res. 10: 1788-1795).
Escherichia coli DB3.1 and DH5α (Invitrogen, Carlsbad, CA, USA) were the hosts for bacterial
manipulations. When non-methylated DNA was needed for enzyme digestions, either GM2163--an E.
coli K12 derivative containing, among others markers, dam13::Tn9 (CamR) and dcm-6 mutations (New
England BioLabs (NEB), Beverly, MA, USA), or JM110--an E. coli K12 derivative containing, among
others, dam and dcm mutations (Yanisch-Perron, et al. 1985 Gene 33: 103-119) was used. E. coli DB3.1
was routinely used to propagate plasmids. In contrast, E. coli DH5α was used only to propagate the
plasmid products of the BP and LR reactions. It is important to understand that Destination Vectors
carrying the ccdB gene cannot propagate in E. coli DH5α and most E. coli strains, because the CcdB
protein, a natural analogue of the quinolone antibiotics (e.g., ciprofloxacin, enoxacin, etc.), binds to the
DNA gyrase subunit A, the product of the gyrA gene, turning it into a cellular poison (Bahassi, et al. 1999
J. Biol. Chem. 274: 10936-10944).
E. coli strains DB3.1 and DH5α were routinely grown in LB liquid culture, or on LB agar plates (15 g/l)
containing the following antibiotics, as indicated in the text: ampicillin (Amp), 150 μg/ml;
chloramphenicol (Cm), 30 μg/ml; kanamycin (Km), 50 μg/ml.
Most DNA manipulations were done following standard procedures as described (Sambrook, et al. 1989,
Ausubel, et al. 1987, Pratt and Aramayo 2002 Fungal Genet. Biol. 37: 56-71). For DNA sequencing we
used the BigDye™ Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase
(PEBiosystems, Foster City, CA, USA). Sequences were generated on an Applied Biosystems Model 377
or 373 automated DNA sequencer at GeneTechnologies Laboratory (Institute of Developmental and
Molecular Biology—IDMB, Texas A&M University, College Station, TX, USA).
PCR reactions were performed in 50 μl reactions for 40 cycles (94°C, 30 s; 60.8°C, 30 s; 68°C, 9.5 min)
with the Clontech Advantage 2 PCR System (BD Biosciences Clontech, Palo Alto, CA, USA) using the
manufacturer’s specifications.
To amplify the 9,001 bp chromosomal DNA region containing the N. crassa NCU02764.1 gene,
we used two primers: OJHAM013 (5'-GGGGACAAGTTTGTACAAAAAAGCAGGCTcgcgaacgtagaaggattattaggcaaagt-3') and OJHAM014
(5'-GGGGACCACTTTGTACAAGAAAGCTGGGTgtcagtcagtcagtcagtcagtcaaccagt-3'). The 29 upper-case characters correspond to the attB1 and attB2 primer sequences present in OJHAM013 and
OJHAM014, respectively. The 30 lower-case characters correspond to the priming sites of the
oligonucleotides in the N. crassa chromosomal region.
After PCR, the resulting PCR products were purified from attB primers and attB primer-dimers by either using PEG precipitation (30% (w/v) PEG 8000/30 mM MgCl2, as recommended by Invitrogen), or extraction from a 1% (w/v) agarose gel and purification with the Wizard PCR Preps DNA Purification Resin as recommended by Promega (Promega, Madison, WI, USA).The LR and the BP GATEWAY™ reactions were both performed as recommended by the manufacturer (Invitrogen).
Results and Discussion
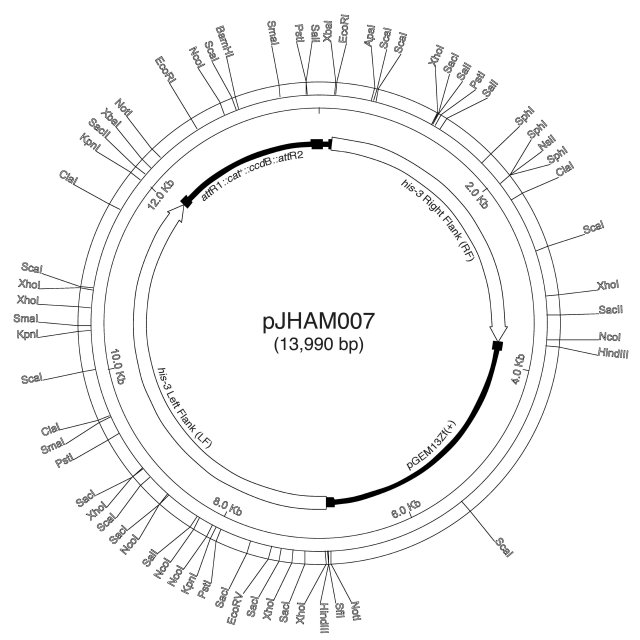
Construction of pJHAM007, a GATEWAY™-compatible his-3-gene replacement Destination Vector. To demonstrate the feasibility of using GATEWAY™ to direct the integration of constructs at the his-3 locus of N. crassa, we started by converting the his-3-gene replacement vector pJHAM003 (Lee, et al. 2003 Curr. Genet. DOI 10.1007/s00294-002-0366-z) into a Destination Vector called pJHAM007. The his-3-gene replacement plasmid, pJHAM007, was constructed by inserting a 2 kb BglII-EcoRV cassette removed from pDEST14 (Invitrogen) and containing the attR1, the cat+ = chloramphenicol acetyl transferase gene (chloramphenicol resistance), the ccdB gene and the attR2, into the BamHI-PmeI restriction sites of pJHAM003 (Figure 1). We confirmed the construction of this plasmid by digesting pJHAM007 using several different restriction enzymes. Following a similar strategy, gene-replacement vectors for other loci can be converted into GATEWAY™ Destination Vectors.
RC-mediated Cloning of PCR Products To Generate Entry Clones. Complementation of a mutant phenotype by an ectopically integrated DNA fragment is a common way to demonstrate gene function. Because filamentous fungi can have large genes and complex promoters, it is not uncommon for those DNA fragments to be large, in order for them to contain all the necessary regulatory elements.
To direct the integration of a DNA fragment at the his-3 locus using the his-3-gene replacement Destination Vector pJHAM007, the PCR fragment first needs to be present in an Entry Clone. It is known that the efficiency of the in vitro RC reactions decreases with increasing size of the DNAs involved (Hartley, et al. 2000 Genome Res. 10: 1788-1795), but given the predicted need to occasionally use large DNA fragments, we first decided to test the limits of the RC-cloning reaction.
We therefore designed PCR primers containing attB sites to amplify a 9 kb DNA region from Linkage Group I (LG I) containing the N. crassa NCU02764.1 gene. PCR products flanked by attB sites can be generated by incorporating attB sites (25 base + 4 G residues) at the 5’-end of PCR primers (attB1 in the forward primer and attB2 in the reverse primer) and cloned by BP Recombination, into attP-containing vectors in the presence of Int and IHF (BP Clonase) to generate Entry Clones capable of recombining with Destination Vectors (Hartley, et al. 2000 Genome Res. 10: 1788-1795). For this, 500 ng of the 9 kb PCR product were mixed with 300 ng of linearized pDONR201 plasmid (Invitrogen) and BP Clonase following the manufacturer’s recommendations (Invitrogen). Following transformation into E. coli DH5α cells and Kanamycin-Resistance (KmR) selection, we analyzed 24 positive clones. Of those, only five were determined to be correct. We selected one correct clone and named it pJHAM009. The resulting plasmid, pJHAM009, is a GATEWAY™-compatible Entry Clone and its insert can therefore be transferred to any other Destination Vector with great versatility and efficiency.
RC-mediated Cloning of the insert from the Entry Clone (pJHAM009), into the his-3-Gene Replacement Destination Vector (pJHAM007), to generate the his-3-Gene Replacement plasmid (pJHAM010). The next step in the construction of a his-3-gene replacement Destination Vector is the transfer of the DNA insert present in the Entry Clone to the Destination Vector. This reaction is catalyzed by the LR Clonase, a mixture of the Int, IHF, and Xis proteins. For this, 140 ng of pJHAM009 Entry Clone were mixed with 300 ng of linearized pJHAM007 plasmid and LR Clonase following the manufacturer’s recommendations (Invitrogen). Following transformation into E. coli DH5α cells and Ampicillin-Resistance (ApR) selection, we analyzed 24 clones, among thousands of transformants. All 24 clones were determined to contain the expected insert size and all had the predicted restriction pattern. We selected one positive clone and named it pJHAM010. These results demonstrate that once in an Entry Clone, inserts as large as 9 kb can be transferred to any other Destination Vector with great versatility and efficiency, especially considering the large size of the resulting plasmid (in this case 21 kb).
Use of plasmid pJHAM007-derivative (pJHAM010) to direct the integration of DNA inserts at the his-3 chromosomal position. To demonstrate the feasibility of using gene replacement plasmids derived from pJHAM007 to direct the integration of DNA inserts at the his-3 chromosomal position, we transformed strains DLNCT62A and DLNCR83A (Lee, et al. 2003 Curr. Genet. DOI 10.1007/s00294-002-0366-z), each using 50 μg of SfiI-linearized plasmid pJHAM010. We selected 12 colonies from each transformation and tested them for FUDR (2’-deoxy-5-fluorouridine or (+)-5-fluorodeoxyuridine, filter sterilized) and hygromycin-resistance/sensitivity as described in (Lee, et al. 2003 Curr. Genet. DOI 10.1007/s00294-002-0366-z). Six FUDR-resistant, hygromycin-sensitive transformants obtained from DLNCT62A and seven obtained from DLNCR83A, were selected for further analysis. DNA was extracted from one DLNCT62A- and one DLNCR83A-derived transformant and Southern blot hybridization analysis was used to determine if a true gene replacement event had occurred in these tranformants. As predicted, both transformants contained the insert present in plasmid pJHAM010 integrated at the his-3 locus (data not shown).
In summary, a large PCR fragment obtained from N. crassa genomic DNA was cloned into a
GATEWAY™-compatible Entry Vector, transferred into a his-3-gene replacement Destination Vector
and integrated back at the N. crassa his-3 chromosomal locus with minimal work and in a short period of
time. We predict that the use of this technology will accelerate the development of our understanding of
N. crassa biology considerably.

Figure 1. pJHAM007, a his-3-gene replacement GATEWAY™-compatible Destination Vector. Circular representation of plasmid pJHAM007 is presented along with the main restriction sites. PGEM13Zf(+) = vector containing a ColE1 origin of replication (Promega), attR1 and attR2 = attachment sites, cat+ = chloramphenicol acetyl transferase gene (chloramphenicol resistance), ccdB = CcdB protein, a quinolone-like antibiotic protein. The left and the right flanks correspond to DNA sequences that flank the site of insertion located downstream of the his-3 gene. The GeneBank Accession number of pJHAM007 is AF541939.
Acknowledgements: This work was supported by U. S. Public Health Service Grant GM58770 to R. A.