Olivera Gavric and Anthony Griffiths, Botany Department, University of British Columbia, Vancouver, Canada V6T 1Z4
Fungal Genet. Newsl. 51: 9-12
A multiple-site molecular dimorphism for the gene for phospholipase C has no known evolutionary basis, yet reveals that Oak Ridge wild types cannot have originated as described in the current best pedigree.
This study concerns nucleotide sequences in wild type alleles of the Neurospora crassa gene for phospholipase C (PLC). We have been interested in this gene regarding its role in tip growth. PLC has been proposed to play a role in calcium signal transduction as follows. Under stretch activation in the plasma membrane, PLC catalyzes the hydrolysis of phosphatidyl inositol 4,5 biphosphate, releasing the second messenger molecules diacylglycerol and inositol-1,4,5-triphosphate (IP3). IP3 binds to a receptor in vacuolar membranes, stimulating the release of Ca++ into the cytosol. Hence PLC plays a key role in maintaining Ca++ gradients in hyphal tip growth (Silvermann-Gavrila and Lew, 2002).
We analyzed the Neurospora crassa strains Oak Ridge 74-OR23-1A (FGSC 987), Oak Ridge 74-OR8-1a (FGSC 988), Abbott 12a (FGSC 1758), Abbott 12a (FGSC 351), Abbott 4A (FGSC 1228), Abbott 4A (FGSC 739), Lindegren 25a (FGSC 353), Lindegren 1A (FGSC 354), Em 5256 (FGSC 424), Em 5297 (FGSC 352), inl a (FGSC 497) and colonial mutant strains col-16a (FGSC 3462), col-16A (FGSC 3461) and col-16a (FGSC 1348). All were obtained from the Fungal Genetics Stock Center. Gene sequences were obtained by PCR amplification. We used primers F-310 (5' CAC TGT ACC GCA CAG CAA GT 3') and R310 (5' TTG CGC ATC GTC TTG TAT TC 3') to amplify the PCR gene (ncu 06245.1) from the different wild type and col-16 strains. The PCR product was cloned into the pCRŽ II-TOPO vector and sequenced. The sequences were compared to ncu 06245.1 in the Neurospora crassa genome sequence (Galagan et al. 2003.).
In the course of routine sequencing we found that the strains show a dimorphism for the gene for PLC. The two morphs, named Morph 1 and Morph 2, differ at 31 nucleotide positions (of which two are in the single intron). Ten missense mutations result. In summary, in this total of 31 changes there is one triplet deletion/addition, 16 silent changes giving the same amino acid (including one case of adjacent changes), and 10 missense changes giving a different amino acid (also including one case of adjacent changes). Of the missense changes, six give amino acids in the same chemical group, and four a different chemical group.
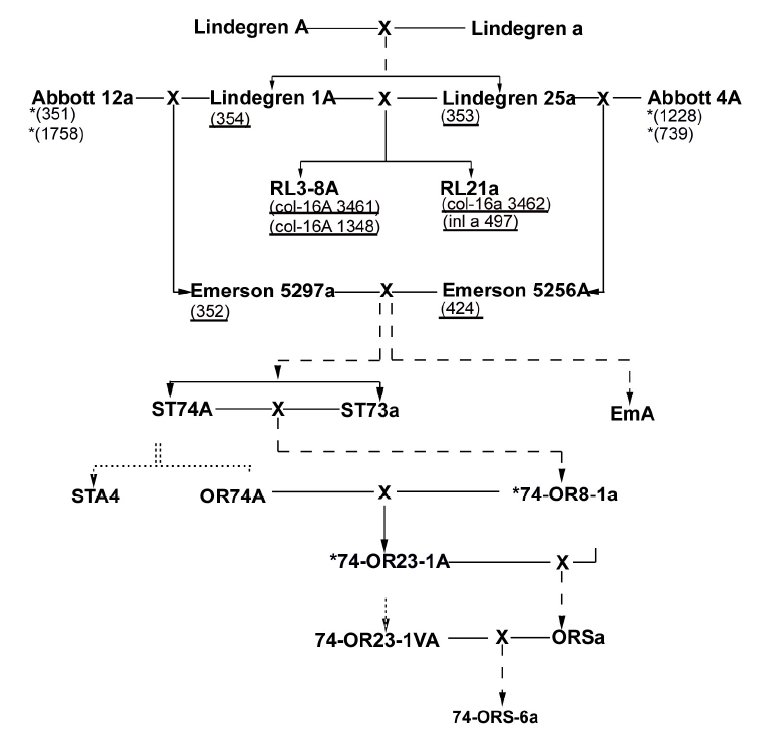
Although the evolutionary significance of this dimorphism is unknown, the distribution of the two morphs of PLC was curious because it is inconsistent with the proposed pedigree of the Oak Ridge wild types. The previously proposed pedigree of the standard Oak Ridge wild type strains is shown in Figure 1. This figure is redrawn from Newmeyer et al. (1987). Those authors considered this pedigree a "best fit" because there were several inconsistencies of the distribution of markers among the strains shown. (The markers are not shown in Figure 1.)
We tested the PLC morphs in several key strains alleged to be ancestors of the Oak Ridge wild types. Both Oak Ridge strains have the same allele, Morph 1. This is expected from inbred strains. However, both Emerson 5297a and Emerson 5256A, which are the supposed direct predecessors of Oak Ridge have Morph 2. Taken at face value, this would mean that the Oak Ridge wild types cannot be descended from the FGSC isolates that allegedly represent Emerson 5297 and 5256. Two Lindegren strains that we have analyzed, 353 and 354, also have the Emerson Morph 2, as do four Rockefeller-Lindegren (RL) strains. However, four Abbott strains designated as 12a or 4A have the Oak Ridge Morph 1, hence the PLC allele of Oak Ridge must be of Abbott ancestry.
The results generally fit the proposed pedigree with the exception of the Oak Ridge branch. With the present data set there is no way to resolve this ancestral inconsistency. The Abbott alleles trickled down through the real pedigree somehow, but apparently not in the way listed in Figure 1. The answer might lie in two possible explanations:
Does the uncertainty about ancestry matter? Probably not from the point of view of the need for a pure reference line, which was the motivation behind Frederick J. deSerres' developing the Oak Ridge stocks. However the results do draw attention to the point that an inbred standard chosen on the basis of asexual and sexual vigor might be individualistic at the level of function. The PLC alleles are a case in point; they have diverged greatly within the sample studied and seem to be potentially different at the functional level. This might reflect their ecological origins. Abbott collected his isolates of N. crassa from Houma, Louisiana, USA (D. D. Perkins, personal communication). Lindegren collected from diverse locations in Cuba, Mexico, Panama and Japan (Lindegren 1934), but the precise origins of the progenitor strains Lindegren A and Lindegren a are not known. Nevertheless the Oak Ridge wild types are clearly fungal melting pots of alleles from diverse ecological settings. The dimorphism raises the question What is a wild type? Even though no-one doubts the need for a standard, to consider that standard as the functional norm is potentially misleading.
References
Galagan, J. E., et al. 2003. The genome of the filamentous fungus Neurospora crassa. Nature 422: 859-868.
Lindegren, C. C., 1934. The genetics of Neurospora V. Self-sterile bisexual heterokaryons. J. Genet. 28: 425-435
Newmeyer, D., D. D. Perkins and E. G. Barry 1987. An annotated pedigree of Neurospora crassa laboratory wild types, showing the probable origin of the nucleolus satellite and showing that certain stocks are not authentic. FGN 34:46-51
Silverman-Gavrila, L.B., and Lew, R.R., 2002. An IP3-activated Ca2+ channel regulates fungal tip growth. J. Cell Science 115 (24), 5013-5025.
Funded by the Natural Sciences and Engineering Research Council of Canada.
Return to the FGN 51 Table of contents
Visit the FGSC homepage