pZHK2, a bi-functional transformation vector, suitable for two step gene targeting
Ulrich Kück and Stefanie Pöggeler.
Lehrstuhl für Allgemeine und Molekulare Botanik, Ruhr-Universität Bochum, 44780 Bochum, Germany
Fungal Genet. Newsl. 51:4 -6
Homologous recombination is a prerequisite for the generation of knock out strains by means of
DNA-mediated transformation. In filamentous fungi however, the frequency of ectopic integration events
is rather high and the actual efficiency of homologous recombination depends upon the length of homologous DNA
flanking the transformation marker. Recently, d´Enfert and coworkers (Chaveroche et al., 2000) presented
a two-step technology for the integration of a bi-functional zeocin-pyrG cassette into a target sequence
of interest using an Escherichia coli strain expressing the phage lambda Red functions. In the resulting
recombinant cosmids, the selection marker is flanked by fungal DNA sequences longer than 1 kb, which can be
used to transform appropriate fungal recipient strains. For selection of fungal transformants, those workers
used the A. nidulans pyrG gene encoding orotidine-5´-monophosphate decarboxylase, which confers
prototrophy in appropriate uridine/uracil auxotrophic recipient strains.
Here, we describe the novel bi-functional transformation vector pZHK2, which carries in addition to the
zeocin resistance gene the hygromycin B phosphotransferase gene often used as a dominant selectable marker
gene in fungal recipient strains. The applicability of the vector is demonstrated by generating a ura3- knock
out strain from Sordaria macrospora showing auxotrophy.
The construction of plasmid pZHK2 was achieved as follows: Initially, we generated plasmid pEM7ZH2 by ligating the 1.4 kb
EcoRI fragment from plasmid pCB1003 (Carroll et al., 1994), carrying the hph gene under the control of the
A. nidulans trpC promoter, into the single EcoRI site of vector pEM7/Zeo (Invitrogen, Carlsbad, CA).
This plasmid harbors the intact ampicillin resistance gene and, consequently, this plasmid can appear as a
contamination in subsequent selection steps in which amplicillin containing media are used for selection
of recombinant clones. Hence, a 1.035 kb BglII-XmnI fragment carrying the kanamycin resistance gene from vector
pCR2.1 (Invitrogen, Carlsbad, CA) was inserted into the XmnI site of pZEM7ZH2. For blunt end ligation, the
5´ protruding BglII site was filled up in a Klenow reaction. This construct has a size of 5301 bp and was
further shortened by deleting a 415 bp BsaI-ScaI fragment. The resulting vector pZHK2 has a size of
4886 bp and is shown in Figure 1.
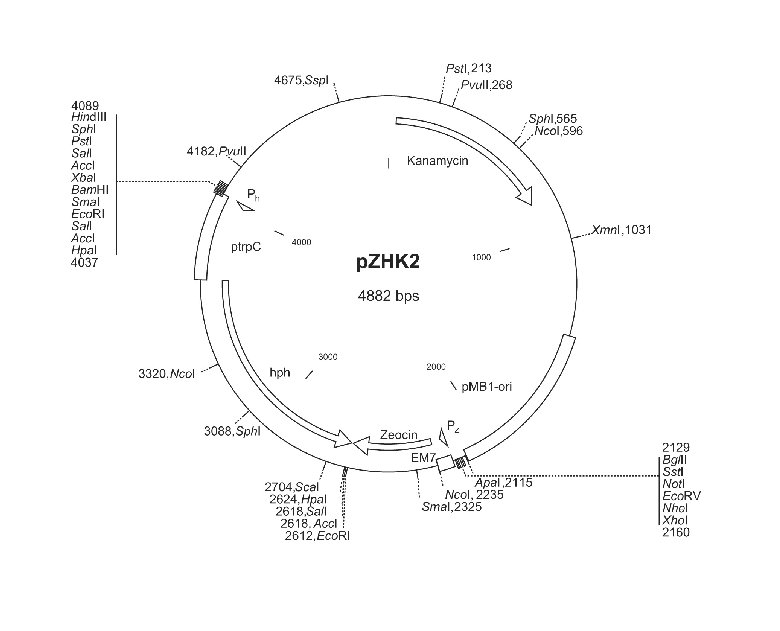
Figure 1: Physical map of the vector pZHK2. Pz and Ph refer to priming sites on the bi-functional zeo-hph cassette. Size
and location of the kanamycin, zeocin and hygromycin B (hph) resistance genes are indicated. The hph gene is fused
with the fungal promoter sequence (ptrpC) from the Aspergillus nidulans trpC gene.
In order to show the applicability of pZHK2, we inserted the zeo-hph cassette in the ura3 gene to generate a uracil
auxotrophic strain from Sordaria macrospora. For this purpose, two 70 bp primers
(Pz-ura3: GACGATGTCGAGCATGCGCGAGAGCTCCTCGCCCTTGCCGACAAGATTGGTGTTGACAATTAATCATCGGCATAG and
Ph-ura3: CCCACCCTGTGATCAGGTCATAGTGGGTCTTGAGGACGACAATCGAGGGGGGGCTTGGCTGGAGCTAGTGGAGG) were synthesized.
The primers have a 50 nt homology to the S. macrospora ura3 gene (Nowrousian and Kück, 1998), followed by 20
nt with homology to the trpC promoter (Ph) or the zeocin resistance gene (Pz). Amplification with
these primers using pZHK2 as a template resulted in a 2 kb fragment containing the zeo-hph cassette that is flanked by
50 bp of the ura3 sequence.
The PCR amplicon was used to transform the E. coli recipient strain KS272 carrying plasmid pKOBEG and plasmid
p20.26 (Chaveroche et al., 2000). The latter contains a 6.6 kb PstI-ClaI restriction fragment with the
S. macrospora ura3 coding region (Fig. 2). After selection of transformed bacteria on LB-medium containing ampicillin and
zeocin, we identified recombinant plasmids in which the zeo-hph cassette was inserted into the ura3 gene. The resulting
plasmid pIG182 (Fig. 2) was used to transform the wild type strain of S. macrospora as described earlier
(Nowrousian et al., 1999).
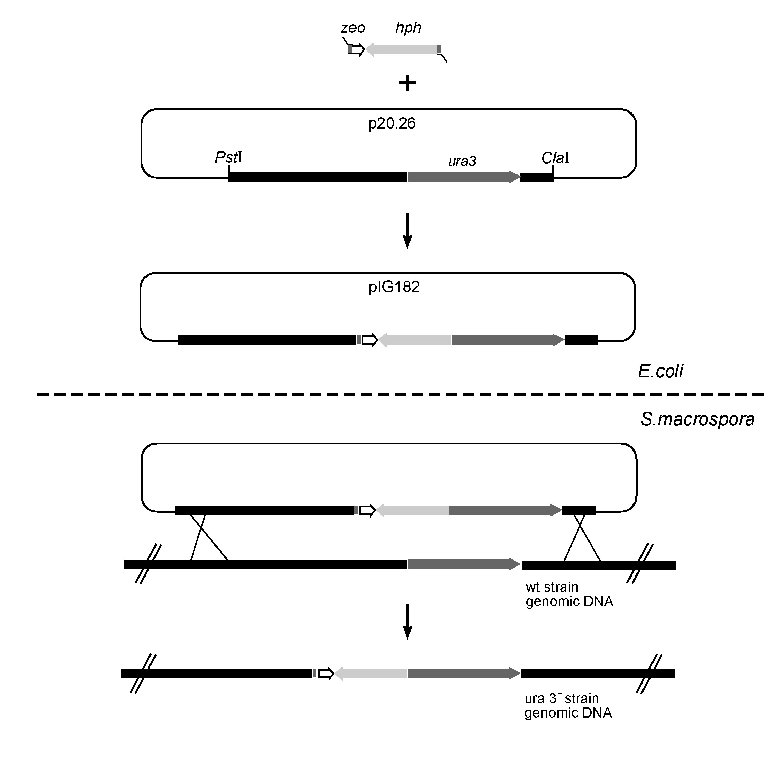
Figure 2: Gene disruption of the ura3 gene in E.coli and in Sordaria macrospora.
Schematic drawing of the disruption construct using pZHK2. The PCR primers shown by arrows were designed
to amplify the region across the zeo-hph cassette and used to screen the mutants carrying the disrupted ura3 gene.
Plasmid p20.26 carries a 6.6 kb PstI-ClaI fragment from the S. macrospora ura3 gene region, and pIG182 is
a derivative, with the zeo-hph cassette inserted into the ura3 coding region (adapted from Chaveroche et al., 2000).
For transformation, plasmid pIG182 was linearized, leaving 5.1 and 1.5 kb of homologous sequence
at each end of the resistance cassette. Fungal transformants were selected on BMM medium supplemented with 100 ug/ml
hygromycin B. A total of 30 fungal transformants were then tested on minimal medium devoid of any supplements. One
transformant, named T7, showed hygromycin B resistance, but was unable to grow on minimal medium without uracil.
Using genomic DNA, a 6.6 kb PstI-ClaI probe detected in a Southern blot a 2.3 kb SacI restriction fragment derived from
the wild type ura3 gene. This fragment can easily be distinguished from the 4.2 kb SacI fragment in T7, which carries
the zeo-hph cassette and is inserted into the ura3 coding sequence. In order to verify the ura3 disruption in
strain T7, plasmid p20.26 was used for DNA-mediated transformation of the auxotrophic recipient strain. The analysis of DNA
from two representative prototrophic fungal transformants is shown in Fig. 3.
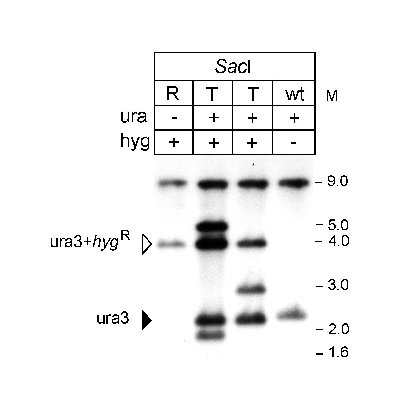
Figure 3: Autoradiograph of a Southern hybridization with genomic DNA from
a S. macrospora wild type (wt) strain, the ura- recipient strain T7 (R)
strain, and prototrophic transformants (T). DNA was treated as indicated and probed with
the radiolabeled PstI-ClaI fragment, shown in Fig. 2.
When compared with the recipient strain,
the transformants showed a complex hybridization pattern, which can be explained by ectopic integration of plasmid
p20.26 into genomic DNA. Using vector pZHK2, we were able to disrupt the ura3 gene, thus generating an auxotrophic
strain, which offers the opportunity to transform S. macrospora with a second marker gene.
In summary, the new bi-functional transformation vector pZHK2 presented here carries a kanamycin resistance marker and
a zeocin-hygromycin resistance cassette, which can be used for in vivo homologous recombinations in E. coli,
S. macrospora and possibly other filamentous fungi. This vector will be useful for transformation of
filamentous fungi showing a low rate of homologous recombination, which can only be transformed with the
hygromycin B resistance marker.
Acknowledgement: We thank Ingeborg Godehardt and Silke Nimtz for excellent technical assistance,
I. Wißmann and E. Szczypka for graphical work, and Dr. Ch. d´Enfert (Paris) for his generous gift
of E. coli strain KS272 (pKOBEG). The work of both authors is supported by research initiative
SFB480 of the ´Deutsche Forschungsgemeinschaft´ (Bonn Bad-Godesberg, Germany).
References
Carroll, A., Sweigard, J. and Valent, B. (1994) Improved vectors for selecting resistance to hygromycin.
Fungal Genet. Newsl., 41, 22.
Chaveroche, M.K., Ghigo, J.M. and d'Enfert, C. (2000) A rapid method for efficient gene replacement in the
filamentous fungus Aspergillus nidulans. Nucleic Acids Res., 28, E97.
Nowrousian, M. and Kück, U. (1998) Isolation and cloning of the Sordaria macrospora ura3 gene and its
heterologous expression in Aspergillus niger. Fungal Genet. Newsl., 45, 34-37.
Nowrousian, M., Masloff, S., Poggeler, S. and Kück, U. (1999) Cell differentiation during sexual development of the fungus Sordaria
macrospora requires ATP citrate lyase activity. Mol. Cell. Biol., 19, 450-460.
Return to the FGN 51 Table of contents
Visit the FGSC homepage


