Xinhua Zhao, Chaoyang Xue, Yangseon Kim, and Jin-Rong Xu. Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN
Fungal Genet Newsl 51:17-18
The conventional approach for generating gene replacement constructs involves several
sequence-specific cloning steps and is time-consuming. A ligation-PCR approach was developed
to efficiently generate gene replacement constructs. Two vectors useful for this ligation-PCR
approach and another vector suitable for improving the efficiency of knockout mutant screens
were constructed.
Flanking sequences longer than 0.5 kb are required to obtain 5-10 % gene replacement transformants in Magnaporthe grisea and many other filamentous fungi. Therefore, the direct PCR approach developed for yeast knockout analysis is not generally applicable. We have used a modified ligation-PCR approach (Lau et al. 2002 J. Microbiol. Meth. 49: 193-205) for generating gene replacement constructs. As outlined in Figure 1, the upstream and downstream flanking sequences of the target gene are amplified with primers 1F and 2R, and 3F and 4R. Primers 2R and 3F contain the AscI and FseI (8-bp cutters) sites, respectively. The resulting PCR products are digested with AscI or FseI and ligated with the hygromycin phosphotransferase (hph) gene released from pCX62 or pCX63 (see below) by AscI and FseI digestion. After ligation, the gene replacement construct can be amplified with primers 1F and 4R or nested primers 5F and 6R.
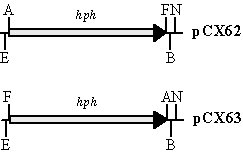
In pCX62, the hph gene is amplified with primers: 5'-AAGAATTCGGCGCGCCGCTGGAGCTAGTGGAGGTCA-3' and 5'-TTGGATCCGGCCGGCCCGGTCGGCATCTACTCTATT-3' from pCB1003 and cloned between the EcoRI and BamHI sites of pBSKS+ (Stratagene). The hph gene also is amplified with primers 5'-AAGAATTCGGCCGGCCGCTGGAGCTAGTGGAGGTCA-3' and 5'-TTGGATCCGGCGCGCCCGGTCGGCATCTACTCTATT-3' and cloned between the EcoRI and BamHI sites of pBSKS+ as pCX63. Both pCX62 and pCX63 have been sequenced and used to transform Magnaporthe grisea and Fusarium graminearum. The hph gene can be released from pCX62 or pCX63 by digestion with FseI and AscI. Another 8-bp cutter enzyme, NotI, can be used to release the hph cassette in the replacement of FseI and AscI from pCX62 and pCX63, respectively.
To increase the ligation efficiency, the hph gene released from pCX62 or pCX63 is dephosphorylated with the shrimp alkaline phosphatase and gel purified. The optimal ratio of the hph fragment and the upstream and downstream flanking sequences is 5: 1: 1. We usually use 25. 5, and 5 ng of the hph gene, upstream, and downstream fragments, respectively, in a 10 l ligation mixture. One microliter of the ligation product is then used in a 50 ul PCR reaction mixture to amplify the gene replacement construct with the Advantage-2 PCR kit (Clontech).
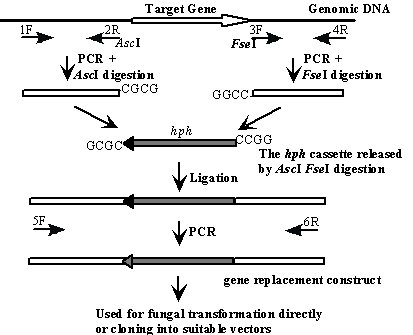
Fig. 1. The ligation-PCR procedure for generating gene replacement constructs
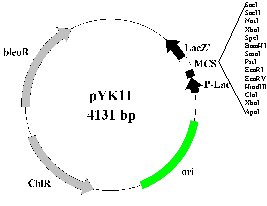
The resulting PCR product can be used directly for fungal transformation or cloned into pYK11 (Fig. 2B) or other suitable vectors. In pYK11, the bleomycin resistance gene (ble) under the TrpC promoter control is cloned in the XmnI site of pBCKS+. Because the SmaI and PstI sites of the ble gene have been removed by site-directed mutagenesis, all the restriction enzyme sites on the MCS of pBCKS+ except SalI can be used for cloning with the blue-white selection. After transformation with gene replacement constructs cloned in pYK11, true knockout mutants will be hygromycin-resistant but bleomycin-sensitive. The ectopic transformants, however, will be resistant to both hygromycin and bleomycin. We have used this ligation-PCR approach to generate gene replacement constructs for six F. graminearum genes and over 20 M. grisea genes.
A. B.
Fig. 2. Vectors pCX62, pCX63, and pYK11. A. The hph gene is amplified and cloned between EcoRI and BamHI sites of pBCKS+ in pCX62 and pCX63. Restriction enzymes are: A, AscI; B, BamHI; E, EcoRI; F, FseI; and N, NotI. B. The modified ble gene is cloned into the XmnI site on pBCKS+. Unique restriction enzyme sites on the multiple cloning site (MCS) of pYK11 are listed.
Return to the FGN 51 Table of contents
Visit the FGSC homepage