Reliable PCR amplification from Neurospora crassa genomic DNA obtained from conidia
Steven T. Henderson, Graham A. Eariss and David E. A. Catcheside – School of Biological Sciences, Flinders University, Bedford Park, SA 5042, Australia.
Fungal Genet. Newsl. 52:24
Boil-mediated lysis of Neurospora conidia (Boil-prep) is an extremely rapid, convenient and useful technique to obtain sufficient genomic DNA template for PCR amplification. We routinely use this technique for screening molecular markers, sequencing, and preliminary confirmation of transformants. However, we have observed periods when successful PCR amplification from Boil-preps has been erratic, hampering the efficacy of this technique. As lysis of conidia results in the liberation of DNA and other cellular components, we reasoned the inconsistent results may have been due to inhibition and/or degradation of the Taq DNA polymerase by cellular material present in the lysis solution. We tried various DNA polymerases and found the reliability of PCR amplification of DNA from Boil-prep template is largely dependant on the specific enzyme used with some Taq polymerases producing <5% successful amplifications. In our hands, Red Hot® DNA polymerase from ABgene (cat#AB-0406) facilitates consistent PCR amplification (>90%) from Boil-prep template DNA. We routinely use Red Hot® DNA polymerase to amplify PCR products up to 1.35kb from Boil-prep DNA following the procedure described below (modified from Yeadon and Catcheside 1996).
Boil-prep method:
A wet loop of 3 - 7 day old conidia is transferred to 100μl of sterile Tris-EDTA (pH 8.0) in a 1.5ml microfuge tube and vortexed briefly. The conidial suspension is placed in a boiling waterbath for 10 minutes and then on ice for 5 minutes. The cellular debris is pelleted by centrifugation at 13000 RPM in a bench-top centrifuge for 5 minutes. 70ul of supernatant is transferred to a new 1.5ml microfuge tube and stored at -20ºC. 2-5ul of Boil-prep DNA solution is used as a template in a 50ul PCR reaction with 0.5U of Red Hot® DNA polymerase.
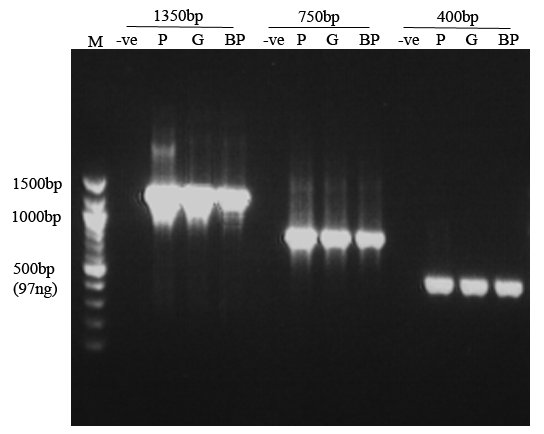
Figure 1. Comparison of PCR amplicons from Boil-prep genomic DNA (BP), genomic DNA (G) (prepared by the method of Irelan et al. 1993), plasmid DNA (P) and no DNA (-ve) templates. Size marker (M) = 100bp DNA ladder (New England Biolabs) with a 500bp reference band that contains 97ng of DNA. Each sample lane contains 6ul of a 50ul PCR reaction. DNA yields from Boil-prep PCR are typically >10ng/ul. Note: all PCRs were performed in a single experiment with cycle conditions optimised for amplification of the 1350bp product.
References:
Irelan, J., V. Miao and E. U. Selker, 1993. Small scale DNA preps for Neurospora crassa. Fungal Genet. Newsl. 40: 24.
Yeadon, P. J., and D. E. A. Catcheside, 1996. Quick method for producing template for PCR from Neurospora cultures. Fungal Genet. Newsl. 43: 71.