Comparison of gpd genes and their protein products in basidiomycetes
Sreedhar Kilaru and Ursula Kües
Molecular Wood Biotechnology, Institute of Forest Botany, Georg-August-University Göttingen, 37077 Göttingen, Germany
Fungal Genetics Newsletter 52:18-23
We compared promoters, coding sequences, introns and terminators of glyceraldehyde 3-phosphate dehydrogenase genes (gpd) from various basidiomycetes. Coding regions of these housekeeping genes are highly conserved (between 60 to 99% DNA identity) whilst non-coding regions have DNA identities of around 40%. Amongst all homobasidiomycete promoters, the TATA region and a CT-rich region with the potential transcription start sites are highest conserved. Surprisingly, there are no other conserved motifs common to all promoters. Up to five introns are clustered at the far 5´ ends of the genes, hinting to a potential function in efficient gene expression.
Little is known about promoters in higher basidiomycetes. Constitutive promoter activities have been described for some homologous and heterologous promoters in Coprinopsis cinerea with the Agaricus bisporus gpdII (glyceraldehyde 3-phosphate dehydrogenase gene 2) promoter being highest in activity (Kilaru et al., 2005). Use of gpd promoters from A. bisporus, Flammulina velutipes, Lentinula edodes, Phanerochaete chrysosporium, Schizophyllum commune and Trametes versicolor has by now been made in different species either for laccase and peroxidase production or for expression of gfp (green fluorescent protein gene) or the bacterial hygromycin resistance gene hph (for references see Kilaru et al., 2005). Surprisingly, homology among these promoter sequences is relatively low (Kilaru et al., 2005). In contrast, the two known gpd genes from A. bisporus (termed gpdI and gpdII), the single gpd gene from P. chrysosporium and an isolated gpd gene from S. commune have been described as highly conserved in intron positions as well as in sequence of their products (Harmsen et al., 1992). Analysis of all gpd genes from basidiomycetes currently present in the NCBI database and of two putative gpd genes of C. cinerea deduced from the published genomic sequence (http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus/) and submitted to the genome annotation database at Duke (http://genome.semo.edu/cgi-bin/gbrowse/coprinus) confirmed this in most other instances (Table 1).
Table 1. Comparisons of sequences from homobasidiomycetous gpd genes
Sequences analyzeda |
Sequence identity/similarity in percentage |
||
Lowest |
Highest |
Mean + standard deviation |
|
Promoterb |
34 |
85 |
41.4 + 5.8 |
Gene (from start to stop codon) |
48 |
78 |
59.3 + 4.8 |
Coding sequencec |
60 (60) |
88 (99g) |
72.0 + 5.2 (72.9 + 5.2) |
Intron 1d |
25 |
57 |
41.0 + 7.5 |
Intron 2d |
23 |
56 |
40.0 + 6.1 |
Intron 3d,e |
22 |
57 |
42.0 + 7.3 |
Intron 4d |
25 |
60 |
40.8 + 7.2 |
Intron 5d |
20 |
54 |
36.5 + 7.8 |
Intron 6d |
24 |
57 |
40.1 + 6.8 |
Intron 7d |
30 |
52 |
41.0 + 5.2 |
Intron 8d |
21 |
61 |
38.5 + 7.5 |
Intron 9d |
32 |
54 |
41.6 + 5.7 |
Terminatorf |
33 |
96 |
40.2 + 9.9 |
Protein productc |
63/77 (63/77) |
88/93 (99/99g) |
74.5 + 4.5/84.8 + 3.2 (75.3 + 4.2/85.0 + 3.1) |
a For source of sequences see Fig. 1, GenBank accession numbers AY842301 and AB075243 for promoters of V. volvacea and T. versicolor and Harmsen et al. (1992) for promoters and introns of A. bisporus, P. chrysosporium and S. commune, respectively.
b 280-300 bp promoter sequence upstream to the startcodon was used except for A. bisporus gpdI, and T. cucumeris where only 264, respectively 200 bp were available.
c Values in brackets include sequences from heterobasidiomycetes.
d The number refers to intron positions in the two A. bisporus genes gpdI and gpdII (Fig. 2).
e Only a conserved 58 bp region was considered from the 121 bp long intron in O. olearius.
f 300 bp sequence downstream to the stopcodon was used except for F. velutipes (113 bp), O. olearius (212 bp) and the unknown basidiomycete (127 bp).
g The high value of 99% from the combination X. dendrorhous and P. rhodozyma has not been included in calculating mean values.
Coding sequences have 60 to 99% DNA identity and the deduced proteins 63 to 99% amino acid identity and 77 to 99% amino acid similarity with the single exception of a Cryptococcus neoformans gpd (gene locus CNI00320). In the best case, its coding sequence has 48% DNA identity to another basidiomycete gene (Thanatephorus cucumeris) and its product 44 % overall identity and 59% overall similarity to another basidiomycete protein (Cryptococcus curvatus). Moreover, it is more closely related to the products of a group of potential gpd genes from ascomycetous species (47% overall identity and 59% overall similarity to Gpd from Aspergillus nidulans gene locus AN2583_2 and 47% overall identity and 60% overall similarity to Gpd from Aspergillus fumigatus gene locus Afu8g02560; Fig. 1). Since the intron distribution in the second putative gpd gene from C. neoformans differs totally from other basidiomycetous genes (Fig. 2) suggesting another, more distant origin, it has been excluded from the further comparisons.
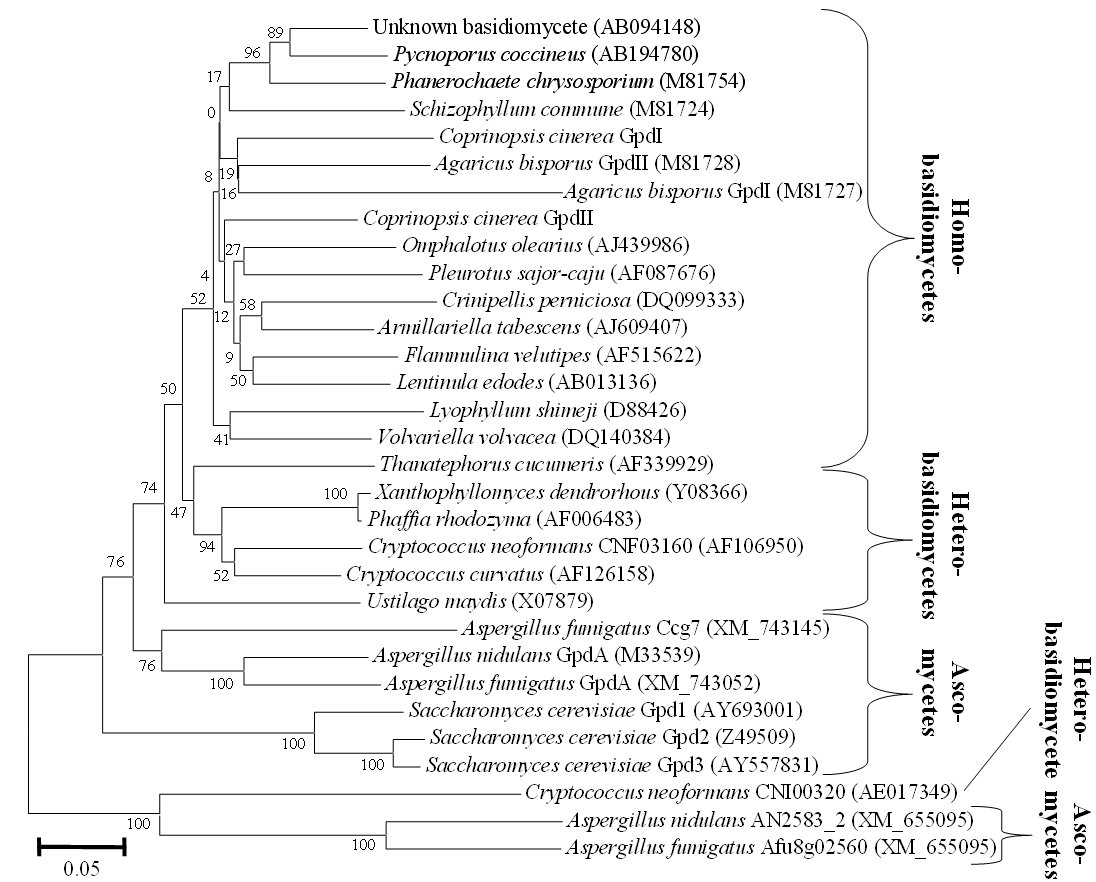
Figure 1. Phylogenetic analysis of protein sequences deduced from (putative) gpd genes from basidiomycetes and a selection of ascomycetes. Two different genes are known in A. bisporus that are localized in tandem in the genome and of which gpdII is functional (Harmsen et al. 1992). Similarly in the C. cinerea genome, there are two genes in tandem that in analogy were called gpdI and gpdII (this study), whilst in the complete P. chrysopsorium genome there is only the one gpd gene originally described by Harmsen et al. (1992). Note that accession number AB094148 refers to C. cinerea strain LT2-44. However, this sequence is likely from another unknown basidiomycete. It has an overall DNA identity of only 64% and 59% to the genes gpdI and gpdII (80% and 70% identity in the coding regions) deduced from the genome of C. cinerea strain Okayama 7 (http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus/). One gpd gene per organism is known from cloning in various other homobasidiomycetes and a few heterobasidiomycetes and one gpd gene has been deduced from the so far incomplete genome of the heterobasidiomycete C. curvatus. The C. neoformans genome contains two different genes (loci CNF03160, CNI00320), the genome of U. maydis one gpd gene, the genome of the ascomycete yeast Saccharomyces cerevisiae three different genes (called gpd1, gpd2, gpd3) and the genomes of the filamentous ascomycetes A. fumigatus and A. nidulans three (gpdA, ccg7 and a gene with no assigned name) and two (gpdA and a gene with no assigned name), respectively.
General structure of gpd genes in basidiomycetes. Between 5 and 10 introns were encountered in the various gpd genes of homobasidiomycetes at mostly conserved positions (corresponding to introns 1 to 9 in the two A. bisporus genes gpdI and gpdII), whilst 1 to 11 introns are present at variable positions in the genes of heterobasidiomycetes (Fig. 2). Only one intron position (intron 3 in A. bisporus genes) is common among all genes but Ustilago maydis. The single intron in the U. maydis gene is however also found in most of the homobasidiomycete genes (intron 2 in A. bisporus genes) but in no other gene of a heterobasidiomycete. Intron 4 in the two A. bisporus genes is conserved in all genes from homobasidiomycetes and in some of the genes from heterobasidiomycetes.
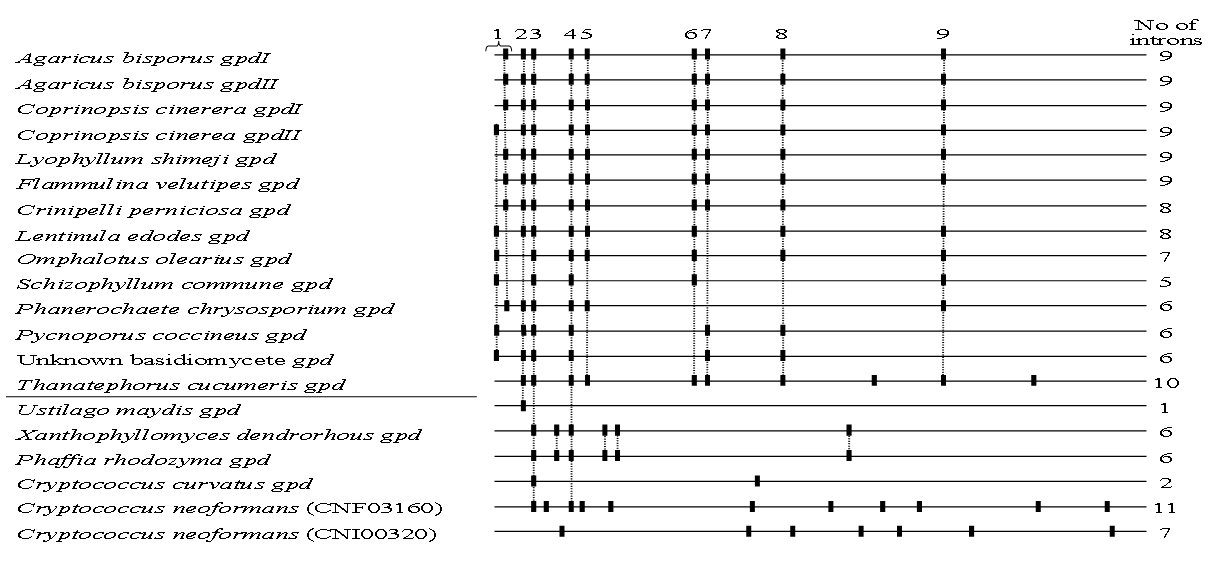
Figure 2. Intron positions within basidiomycete gpd genes. Intron positions for A. bisporus gpdI and gpdII (labelled with numbers 1 to 9) and the gpd genes from P. chrysosporium and S. commune were deduced from Harmsen et al. (1992). All others were from sequences in the NCBI GenBank (for accession numbers see Fig. 1). A horizontal line between species names separates genes from homo- and heterobasidiomycetes. The } under the number 1 indicates an intron position that due to insertions or deletions of one or two codons 3´ to the ATG start-codons are at seemingly different locations.
Striking is the observation that the majority of introns (including the three conserved between homo- and heterobasidiomycetes) localize in the 5´ half of the genes whilst in the 3´ half most genes carry no or only one intron. Within 30 bp at the 5´ end of all genes are up to three introns and, in most cases, up to four more in the next 300 bp of coding sequence. Nine of the homobasidiomycetes have the first intron inserted directly after the ATG start-codon. Insertion or deletion of one to two codons 3´of the startcodon in individual genes likely moved this first intron into two seemingly different spatial positions (labeled intron 1 in Fig. 2). The presence of the first intron after the startcodon in the A. bisporus gene allowed expression of the gfp (green fluorescent protein) gene from the gpdII promoter in hyphae of A. bisporus, C. cinerea and P. chrysosporium (Ma et al., 2001; Burns et al., 2004) whilst GFP was not detected in transformants of homobasidiomycetes when this intron was lacking (Chen et al., 2000; Godia et al., 2004). It is possible, that this first intron functions as well in efficient gpd expression. Likewise, the accumulation of several introns at the beginning of the coding regions might serve better gene expression. This assumption is of importance when using the fungi for protein production and needs to be tested in the future.
Sequence analysis in homobasidiomycetes. Coding sequences among genes from homobasidiomycetes are highly conserved (in average 72.0 + 5.2% DNA identity), unlike promoters, introns and terminators that have, on average, around 40% DNA identity (Table 1). A phylogenetic analysis of all introns from homobasidiomycetes showed that neither introns within a given gene nor introns at a given position tended to be more conserved to each other than to introns of other genes at other positions (not shown). The introns had a typical basidiomycete length of 45-84 bp (except the 121 bp long Omphalatus olearius sequence at intron position 3), the canonical GT and AG splice junctions and in 96 of 109 total cases the internal CTNA consensus sequence for lariat formation (Hoegger et al., 2004).
Promoters of gpd genes are expected to be highly active during vegetative growth (Hirano et al., 1999). Promoter activity of the A. bisporus gpdII genes resides within 265 bp upstream to the gpd startcodon (van de Rhee et al., 1996) and of the S. commune gpd gene within only 130 bp (Schuren and Wessels, 1994). In the first 300 bp of the promoter regions of homobasidiomycete genes, only L. edodes and Volvariella volvacea (85% DNA identity), respectively T. versicolor and an unknown basidiomycete (61% DNA identity) showed significant homology. Between any other two given promoter sequences, there are only short conserved DNA stretches which however differed between the combinations of promoters looked at. Nevertheless, common motifs in promoters might still be expected for the different species. The highest similarity amongst all promoter sequences is found in the first 100 bp upstream of the startcodon, covering a CT-rich region and a TATA-motif. Predicted transcription start sites correlate with the CT-rich regions (Fig. 3) and perfectly or nearly perfectly with experimentally confirmed transcription start sites (Harmsen et al., 1992; Hirano et al., 1999). No transcription initiation site was prediced for A. bisporus gpdI, that is not transcribed (Harmsen et al. 1992) and for C. cinerea gpdI, that locates in tandem upstream to the gpdII gene in a same pattern as the two genes in A. bisporus (Harmsen et al., 1992).
Only in exceptional cases, a CAAT-motif is present in the promoter sequences (Fig. 3). No other patterns common to all promoters were identified by the MEME program (http://meme.sdsc.edu/meme/meme.html). Searches in the Transfac database with Motif Search (http://motif.genome.jp/) and TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) found in some promoters a potential binding site for a StuA-type transcription factor and mostly also one or two potential NIT2 binding sequences, but these at non-conserved positions (Fig. 3). Otherwise, in most promoter sequences are at non-conserved places some heat shock elements and potential ADR1 and GATA-factor binding sites (not shown). Using longer sequences where available (up to 2170 bp), searches did not reveal further conserved elements between all the promoters.

Figure 3. Promoter alignments (up to 300 bp sequence upstream to the startcodon). Predicted transcription start points (http://www.fruitfly.org/seq_tools/promoter.html) within CT-rich regions (hand adjusted), TATA regions (hand adjusted), CAAT motifs, StuA-like binding motifs (NWWCGCGWNM) and potential NIT2 sites (TATCTM) are boxed. The three-letter-code indicates the species a gene comes from (first letter from genus, the other two from species name). The Roman number refers to the gene number in case a species has two different genes.
Terminator sequences were also observed in a multiple alignment (not shown). All had a sequence T100%G100%T91%A/G81%A/G72%T/C63%A/G91%A/G81%A/G81%T/C91%A/G81%NA/C91%C/T81%X1-4T100% positioned 75 to 150 bp downstream to the stop codon. In L. edodes, this is 3 and 40 bp, respectively upstream to the two known poly-A sites (Hirano et al. 1999). In S. commune, an mRNA 3´ end has been localized within this sequence (Harmsen et al., 1992).
Sequence analysis in heterobasidiomycetes. The overall levels of sequence conservation amongst genes from heterobasidiomycetes are similar as in homobasidiomycetes. Coding sequences of genes from heterobasidiomycetes have in average 76.7 + 2.6% identities to each other and the deduced proteins identities of 79.6 + 4.1 and similarities of 88 + 2.6% [excluding the combination Xanthophyllomyces dendrorhous and Phaffia rhodozyma originally thought to represent one species (Fell and Blatt, 1992)]. Promoter sequences (300 bp) have an average DNA identity of 39.3 + 4.3% and terminator sequences an average identity of 38.0 + 3.5%. The only prominent features within the promoter sequences are CT-rich stretches but without a clear TATA-element (not shown). There were no other motifs from the TransFac database common between the four analyzed promoters. Within 300 bp terminator sequences there was no conserved sequence as in the terminator regions of the homobasidiomycete genes (not shown).
Conclusions. Coding sequences among constitutively active gpd genes in basidiomycetes are highly conserved in contrast to non-coding promoters, introns and terminators. The strong promoters of homobasidiomycetes share surprisingly few promoter elements, mainly a TATA motif and a downstream CT-rich stretch that seem to define the transcription initiation start site. In the heterobasidiomycetes, there appears to be only the CT-rich stretch. Otherwise, the lack of potential regulatory elements raises the idea that highly active constitutive promoters may function rather by absence of transcription factor binding sites. A 130 bp fragment of the S. commune gpd promoter containing little more than the TATA-sequence and the CT-rich stretch enabled expression of a bacterial ble gene for phleomycin resistance in the fungus (Schuren and Wessels, 1994). In L. edodes, a homologous 329 bp promoter fragment was ineffective in hph expression in contrast to a larger 511 bp fragment (Hirano et al., 2000). However, as shown in studies with the constitutive active tub1 promoter in C. cinerea, a larger promoter fragment does not necessarily function better than a shorter one (Burns et al., 2005; Kilaru et al., 2005). A. bisporus gpdII promoter fragments 265 and 277 bp in length have been shown to be sufficient for function in A. bisporus and C. cinerea. Even so, intron sequences were sometimes needed for successful gene expression (van de Rhee et al., 1996; Burns et al., 2005; Kilaru et al., 2005). In the S. commune and L. edodes studies mentioned above, introns within the genes were lacking (Schuren and Wessels, 1994; Hirano et al., 2000) but expression of gfp and various hydrophobin genes in S. commune required an intron for transcript stabilization (Lugones et al., 1999) and presence of an intron also supported hph expression (Scholtmeijer et al., 2001). In the study by Lugones et al. (1999), transcription was not enhanced by presence of the intron localized at the 3´ end of hydrophobin gene SC6. Still, the accumulation of introns at the 5´ end of the gpd genes might positively influence transcription initiation next to transcript stabilization. The currently available data do not allow a final conclusion on what is required in basidiomycetes for highly efficient gene expression.
Acknowledgment. Our laboratory is supported by the Deutsche Bundesstiftung Umwelt (DBU).
References
Burns, C., K.E. Gregory, M. Kirby, M.K. Cheung, M. Riquelme, T.J. Elliott, M.P. Challen, A. Bailey, and G.D. Foster. 2005. Efficient GFP expression in the mushroom Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet. Biol. 42: 191-199.
Chen, X., M. Stone, C. Schlagnhaufer, and C.P. Romaine. 2000. A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl. Environ. Microbiol. 66: 4510-4513.
Fell, J.W., and G.M. Blatt. 1999. Separation of strains of the yeasts Xanthophyllomyces dendrorhous and Phaffia rhodozyma based on rDNA IGS and ITS sequence analysis. J. Ind. Microbiol. Biotechnol. 23: 677-681.
Godia, R.P., R. Fouces, E.J. Gudiña, and J.F. Martin. 2004. Agrobacterium tumefaciens-mediated transformation of the antitumor clavaric acid-producing basidiomycete Hypholoma sublateritium. Curr. Genet. 46: 287-294.
Harmsen, M.C., F.H.J. Schuren, S.M. Moukha, C. M. Vanzuilen, P.J. Punt, and J.G.H. Wessels. 1992. Sequence analysis of the glyceraldehyde-3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr. Genet. 22: 447-454.
Hirano, T., T. Sato, K. Okawa, K. Kanda, K. Yaegashi, and H. Enei. 1999. Isolation and characterization of the glyceraldehydes-3-phosphate dehydrogenase gene of Lentinus edodes. Biosci. Biotechnol. Biochem. 63: 1223-1227.
Hirano, T., T. Sato, K. Yaegashi, and H. Enei. 2000. Efficient transformation of the edible basidiomycete Lentinus edodes using a vector using a glyceraldehyde-3-phosphate dehydrogenase promoter to hygromycin B resistance. Mol. Gen. Genet. 263: 1047-1052.
Hoegger, P.J., M. Navarro-González, S. Kilaru, M. Hoffmann, E.D. Westbrook, and U. Kües. 2004. The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45: 9-18.
Kilaru, S., P.J. Hoegger, A. Majcherczyk, C. Burns, K. Shishido, A. Bailey, G.D. Foster, and U. Kües. 2005. Expression of laccase gene lcc1 in Coprinopsis cinerea under control of various basidiomycete promoters. Appl. Microbiol. Biotechnol. DOI: 10.1007/s00253-005-0128-1.
Lugones, L.G., K. Scholtmeijer, R. Klootwijk, and J.G.H. Wessels. 1999. Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol. Microbiol. 32: 681-689.
Ma, B., M.B. Mayfield, and M.H. Gold. 2001. The green fluorescent protein gene functions as a reporter gene expression in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 67: 948-955.
Scholtmeijer, K., H.A.B. Wösten, J. Springer, and J.G.H. Wessels. 2001. Effect of introns and AT-rich sequences on expression of the bacterial hydrophobin B resistance gene in the basidiomycete Schizophyllum commune. Appl. Environ. Microbiol. 67: 481-483.
Schuren, F.H.J., and J.G.H. Wessels. 1994. Highly efficient transformation of the homobasidiomycete Schizophyllum commune to phleomycin resistance. Curr. Genet. 26: 179-183.
van de Rhee M.D., P.M.A. Graca, H.J. Huizing, and H. Mooibroek. 1996. Transformation of the cultivated mushroom, Agaricus bisporus, to hygromycin B resistance. Mol. Gen. Genet. 250: 252-258.