A ras-1bd Mauriceville strain for mapping mutations in Oak Ridge ras-1bd strains
A. Kaitlyn Beasley*, Teresa M. Lamb*, Wayne K. Versaw and Deborah Bell-Pedersen*
*Center for Biological Clocks Research and Department of Biology, Texas A&M University, College Station, TX 77843
Fungal Genetics Newsletter 53:30-33
We describe the construction of a Neurospora crassa Mauriceville strain carrying the ras-1bd mutation marked by the bacterial hygromycin resistance gene, hph (new FGSC # 10156). This strain is valuable for mapping mutations in Oak Ridge strains that carry the bd mutation.
The bd mutation has been a benefit to circadian rhythm research as it slows down wild type growth rates and sharpens the conidial banding pattern on racetubes (Sargent et al., 1966; Bell-Pedersen et al., 2005). Recent work (Belden et al., 2006) has shown that the bd mutation lies in the ras-1 gene. Mutations that affect circadian banding patterns are typically isolated in strains carrying the ras-1bd allele. Mapping such mutations by the CAPS method (Jin et al., in press) requires that the mutant phenotype be followed in a cross to a wild type Mauriceville strain. Assuming non-linkage, only half of the progeny will be bd, and thus readily scorable. Insertion of the ras-1bd allele into the Mauriceville parent renders all progeny useful for segregation analysis. Furthermore, marking ras-1bd with the hygromycin resistance gene, hph, permits a simple drug resistance test to determine linkage to ras-1.
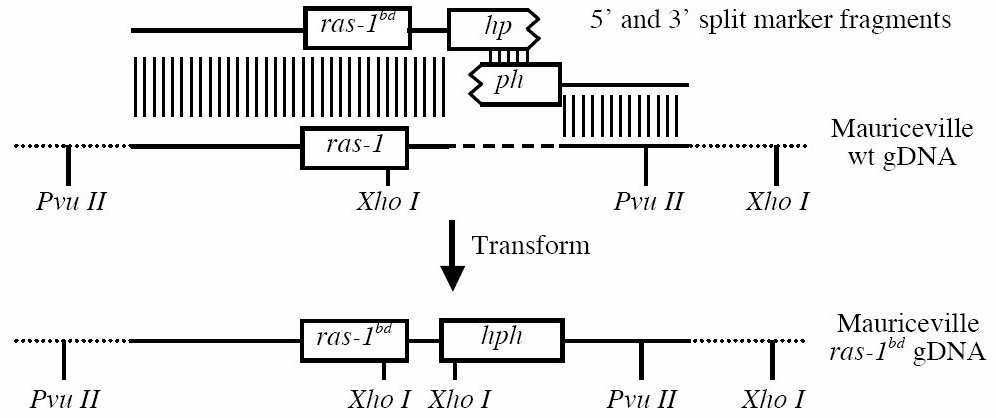
The strategy for gene replacement was similar to published split marker gene deletion strategies (Catlett et al., 2003; Colot et al., 2006). However, rather than deleting the wild type gene, we replaced it with the bd allele and inserted the hph gene in the 3'UTR at position + 225 from the stop codon (Figure 1).

Figure 1. Schematic diagram of ras-1 genetic manipulations. Regions of homology are indicated by vertical hatch marks; there are 763 bp of homology between the split hph fragments, 3223 bp of homology at the 5' end, and 2582 bp of homology at the 3' end. Pvu II and Xho I sites are marked to show the differences between the wild type (wt) genomic (g) locus and the proper ras-1bd-hph integrant. The dashed line indicates a gap where the hph gene is integrated and the dotted lines indicate flanking genomic DNA that remains unchanged.
The replacement fragments were generated in two steps. The first step involved amplification of ras-1bd genomic regions from the Oak Ridge bd strain (FGSC 1858). The 5' region was amplified using primers ras1 F1 and ras1-hyg R1, while the 3' region was amplified using primers hyg-ras1 F2 and ras1 R2. The second step was to generate hybrid pieces that contained a portion of the bacterial hph gene fused to either 5' or 3' regions of ras-1bd (Figure 1). The 5' split marker fragment was generated by amplification of the 5' region of ras-1bd and the complete hph gene, with primers ras1 F1 and YG2. The 3' split marker fragment was generated by amplification of the 3' region of ras-1bd and the complete hph gene, with primers HY2 and ras1 R2. The complete hph gene was obtained by amplification of pBP15 (a gift from D. Ebbole) with primers M13F2 and M13R2. The pBP15 plasmid contains a 1.4kb HpaI PtrpC-hph fragment from pCB1003 (Carroll et al., 1994) subcloned into the EcoRV site of pBluescript II SK (Stratagene, TX). The sequences for all primers used are shown in Table 1, and the genotypes of strains are listed in Table 2.
Table 1
Name of primer |
Sequence (5' to 3') |
ras1 F1 |
TGCATGCTGTGCCGCCTAAC |
ras1-hyg R1* |
actggccgtcgttttacaacgtcgtgacTAAGGTATGGGCGTGGCCGT |
hyg-ras1 F2* |
ggtcatagctgtttcctgtgtgaaattgGCTCACGTACGGTGACCAGT |
ras1 R2 |
TATGGTGGAGCTACCGACAG |
HY2 |
gttggtcaagaccaatgcggagca |
YG2 |
cgacagcgtctccgacctgatg |
M13F2 |
gtcacgacgttgtaaaacgacggccagt |
M13R2 |
caatttcacacaggaaacagctatgacc |
* lower case sequence corresponds to hph homology regions.
Table 2
Name of strain |
Genotype |
FGSC 1858 |
Oak Ridge ras-1bd mat A+ |
FGSC 2225 |
Mauriceville-1c mat A+ |
FGSC 10156 (FGSC 2225 tfm10) |
Mauriceville-1c ras-1bd mat A+ |
The 5' and 3' split marker products were co-transformed by electroporation into the wild type Mauriceville-1-c mat A+ strain (FGSC 2225) (Colot et al., 2006). We restricted this work to the mat A+ strain since the mat a+ Mauriceville-1d strain (FGSC 2226) cannot be used as a crossing partner with an Oak Ridge strain of opposite mating type for CAPS mapping. This is because the Mauriceville-1d strain is an independent isolate that possesses primarily Oak Ridge-type polymorphisms (Jin et al., in press). Several (~50) hygromycin resistant colonies grew, and conidia from 12 of these were extruded through a 5µm filter (Millex-SV, Millipore) to obtain homokaryotic micro-conidia (Ebbole and Sachs, 1990). Each isolate was placed on racetubes for growth rate determination and period determination of the conidiation rhythm. Seven out of twelve transformants had growth rates and periods consistent with replacement of the wild type ras-1 gene with the bd allele; growth rates were reduced from the wild type Mauriceville level of ~7.8 cm/day to ~2.7 cm/day, and their conidial bands became more focused, permitting period measurement (~22.4 hours). One of these transformants (tfm10) was grown and conidia were extruded through a 5µm filter a second time to ensure isolation of a homokaryon. Figure 2 shows the race tube phenotype of the doubly purified tfm10 compared to Oak Ridge bd (FGSC1858) and Mauriceville wild type (FGSC 2225) strains. Thus, tfm10 is hygromycin resistant and carries the bd allele of ras-1.
To verify that recombination occurred as expected at the ras-1 locus, and that extra copies of the inserted DNA were not present, we performed Southern blot analysis on the doubly purified tfm10 (Figure 3). As expected, digestion with Pvu II and hybridization with a 5' ras-1 probe yielded a 5144 bp signal in the wild type Mauriceville strain (FGSC 2225), and a larger 6804 bp signal in tfm10. Digestion with Xho I and hybridization with a hph probe yielded no signal in the wild type Mauriceville strain, and a unique 5655 bp signal in tfm10. Thus, the doubly filtered tfm10 has recombined without additional ectopic insertion. We have deposited this transformant to the Fungal Genetics Stock Center, Neurospora strain FGSC 10156.

Figure 2. Racetube phenotype. Racetubes containing 1 x Vogels Medium N, 0.1% glucose, 0.17% arginine, and 1.5% agar were inoculated with conidia from the indicated strains and grown for one day at 30°C in the light. The position of the growth front was marked, and tubes were then shifted to constant darkness at 25°C. The growth front was marked subsequently every 24 hours (vertical black lines). Controls include the wild type Mauriceville strain (FGSC 2225) and an Oak Ridge bd strain (FGSC 1858).
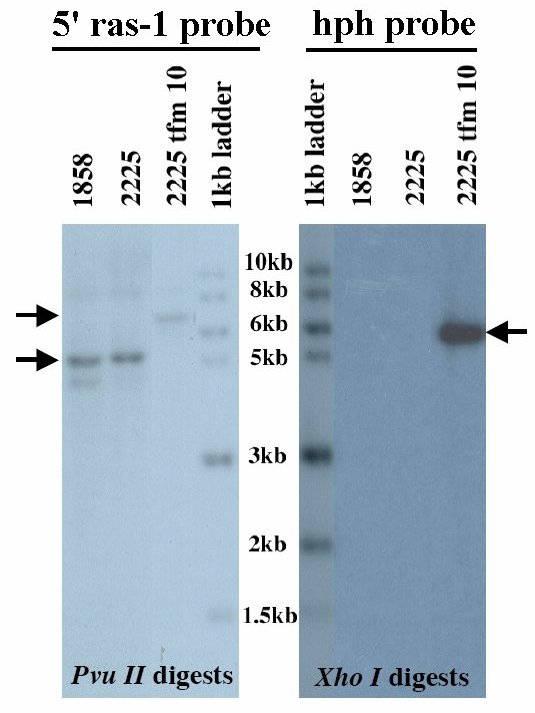
Figure 3. Southern blot analysis of Neurospora strains. Genomic DNA from the indicated strains was digested with Pvu II and probed with a labeled 5' ras-1 fragment or digested with Xho I and probed with a labeled hph fragment. Blots were hybridized overnight, washed, and exposed to film for 16 hours.
We are currently using this new ras-1bd Mauriceville strain to genetically map a mutation isolated in the Oakridge bd background that permits circadian conidiation rhythms to occur in constant light. This strain will also permit rapid determination of linkage to ras-1 using a simple hygromycin resistance test, and will allow mapping of any mutation in Oak Ridge strains that depends on the ras-1bd allele. Investigators studying genetic interactions with the RAS pathway may find this strain useful. Lastly, the methods described to create the knock-in ras-1bd allele are applicable for generating strains carrying point mutations in any Neurospora gene of interest, and in particular for investigating the function of essential genes.
References:
Belden, B., L. Larrondo, M. Shi, A. Froehlich, J. J. Loros, and J. C. Dunlap, 2006. Genetic and molecular characterization of the Neurospora crassa bd mutation: The molecular basis of the overt circadian rhythms in Neurospora. Abstract: Tenth Meeting Society for Research on Biological Rhythms.
Bell-Pedersen D., V. M. Cassone, D. J. Earnest, S. S. Golden, P. E. Hardin, T. L. Thomas, and M. J. Zoran, 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6(7): 544-556.
Carroll, A. M., J. A. Sweigard and B. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41: 22.
Catlett, N. L., B. Lee, O. C. Yoder, and B. G. Turgeon, 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50: 9-11.
Colot H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap, 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103(27): 10352-10357.
Ebbole, D. and M. S. Sachs, 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37: 17-18.
Jin, Y., S. Allan, L. Baber, E. K. Bhattarai, T. M. Lamb and W. K. Versaw, Rapid genetic mapping in Neurospora crassa. Fungal Genet. Biol. In press.
Sargent, M. L., W. R. Briggs and D. O. Woodward, 1966. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 41: 1343-1349.
Return to the FGN 53 Table of Contents