Neurospora msh-4 ortholog confirmed by split-marker deletion
Sue Conway, Fred Bowring, Jane Yeadon and David Catcheside
School of Biological Sciences, Flinders University, PO Box 2100, Adelaide, South
Australia 5001
Fungal Genetics Newsletter 53:5-8
Although most eukaryotes have both MSH4 and MSH5 orthologs, Neurospora was initially thought to lack msh-4. We have deleted the most likely msh-4 candidate and observed a delay in the sexual cycle, disruption to meiosis and a reduction in fertility. Deletion is dominant, showing msh-4 is subject to MSUD. We conclude that Neurospora has a MSH4 ortholog and that it may have remained undetected because of an unusually high number of introns.
MSH4, a member of the MutS protein family, is conserved in higher organisms and has a role in crossover regulation. Disruption of MSH4 reduces the frequency of crossing over in many organisms (Ross-Macdonald and Roeder, 1994; Zalevsky et al., 1999) and increases the incidence of chromosomal non-disjunction, identical phenotypes to that of MSH5 mutants (Hollingsworth et al., 1995; Hunter et al., 1997). MSH4 and MSH5 function as a heterodimer, and removal of the ATPase domain from either leads to loss of function (Alani, 1997; Haber and Walker, 1991; Pochart et al., 1997). All MSH4 orthologs contain MutS DNA-binding and ATPase domains. MSH4 also seems to have a role in the formation or elongation of the synaptonemal complex, a proteinaceous structure that holds homologous chromosomes in close proximity prior to the reductional division of meiosis. Chromosome synapsis is delayed and incomplete in Saccharomyces cerevisiae MSH4 mutants (Novak et al., 2001), and MSH4 localises at synapsis initiation sites in both yeast and mammalian meiotic cells (Novak et al., 2001; Neyton et al., 2004).
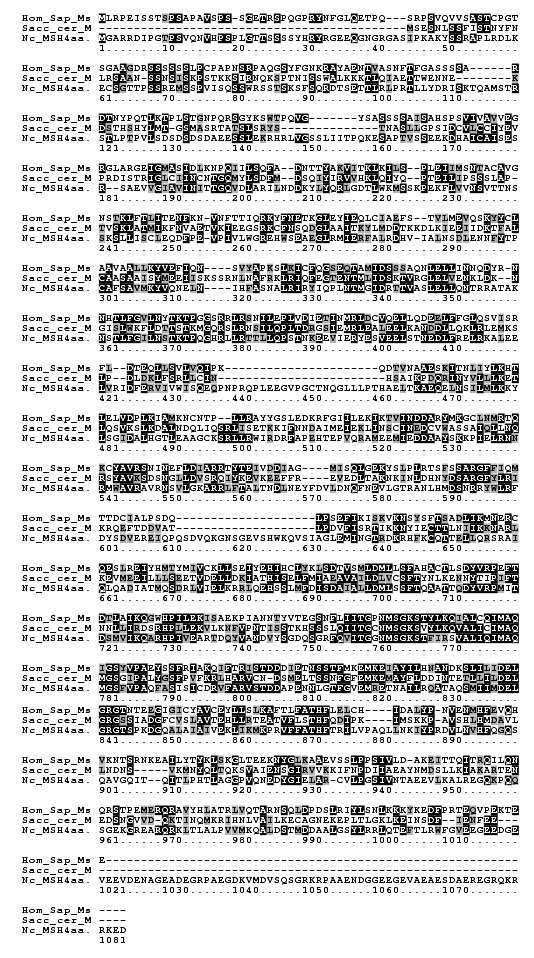
Gene prediction algorithms used in the Neurospora genome project initially failed to identify a MSH4 ortholog. A subsequent bioinformatic analysis used tBlastn to identify a region with homology to MSH4 sequences of S. cerevisiae, Mouse and Human (Borkovich et al., 2004). Alignment of the predicted protein sequence with human and S. cerevisiae MSH4 protein sequences using ClustalW showed that Neurospora MSH-4 shares conserved regions with the other two amino acid sequences (Figure 1).
We concluded that the msh-4 gene is about 3.5 kb in length (Figure 2), and is located on LG1R, between met-6 and aap-2, on supercontig 7.2, nucleotide positions 1061364-1064888. This predicted mRNA includes most of the Neurospora predicted gene NCU10895.2, although it lacks 825 bp at the 5´ end. We decided on the amino terminal of the MSH-4 protein based on homology to other sequences (Figure 1) and although the amino terminal of NCU10895.2 has no homology to other MSH4 proteins, without cDNA information to support our annotation, it is possible that this section is also part of the Neurospora MSH-4 protein.
A 3.8 kb sequence including the potential msh-4 gene was analysed using GenScan (Burge and Karlin, 1997) to identify possible coding sequence and to suggest possible amino acid sequences by removing introns predicted by identification of eukaryotic splice sites (Figure 2). Using either human or Arabidopsis parameters gave very similar results.
Analysis of the predicted 1073 amino acid sequence using Motifs (GCG: Wisconsin PackageTM) identified a potential gene with an ATP/GTP binding site and a DNA MMR motif. Use of a CD search, which identifies conserved domains within a protein sequence by comparing the sequence with other known protein sequences (Marchler-Bauer et al. 2005), identified a MutS homolog with a DNA binding mismatch repair domain (MUTSd) and an ATPase domain (MUTSac; figure 3).
Thus, MSH-4 appears to possess DNA-binding and ATPase domains as expected, and it seems likely that there are six introns (Figure 2) within the msh-4 nucleotide sequence. Since Neurospora genes have, on average, 1.7 introns (Borkovich et al., 2004), the unusually large number in this msh-4 candidate might explain why it was originally missed by the gene prediction algorithms.
Using split-marker deletion (Catlett et al., 2003), we replaced the predicted msh-4 sequence, including all of NCU10895.2, with the hygromycin resistance gene (hph) in a mating pair of Neurospora strains, and analysed the effect through comparison of crosses that are isogenic except for the msh-4 deletion. Crosses were initiated by co-inoculation onto crossing medium and perithecia were examined when mature asci were visible, approximately 13 days after inoculation.

Figure 1. Alignment of the human and S. cerevisiae MSH4 proteins with the Neurospora predicted 1073 amino acid sequence. Dark shading represents identical and lighter shading similar residues.
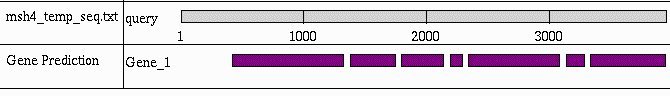
Figure 2. GenScan graphical representation of predicted coding sequences within the putative msh-4 nucleotide sequence. The gaps between blocks represent predicted introns.
Gross perithecial morphology seems to be unaffected by the deletion, although ejection of the first spores is delayed by ~2 days in mutant crosses. There is a 5-fold reduction in viable spores recovered from Dmsh-4 homo- and hetero-zygotes (mutant average = 2 x 105 spores per cross, untransformed control = 1 x 106 spores per cross) and mutant crosses yield many more white spores. Sporogenesis in homozygous and heterozygous Dmsh-4 crosses is delayed and rosettes from the Dmsh-4 mutants contain more bubble asci (Perkins and Barry, 1977) and fewer normal size asci than the control. Asci containing less than eight spores or at least one misshapen spore are common in Dmsh-4 crosses but rarely observed in the control (figure 4).
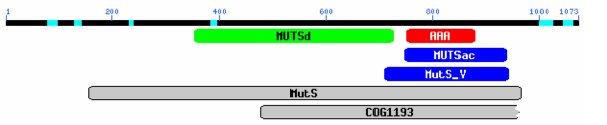
Figure 3. CD graphical representation of conserved domains within the MSH-4 predicted protein sequence. MutS (MutS ATPase protein family), MutSd (DNA binding domain), MutSac (ATPase domain), MutS_V (domain V; ATPase). Gaps represent sequences with low homology to known protein domains
A cursory analysis of chromosomal behaviour during meiosis suggests that an absence of msh-4 causes some abnormality, although at this stage we have not found evidence of non-disjunction during meiosis I.
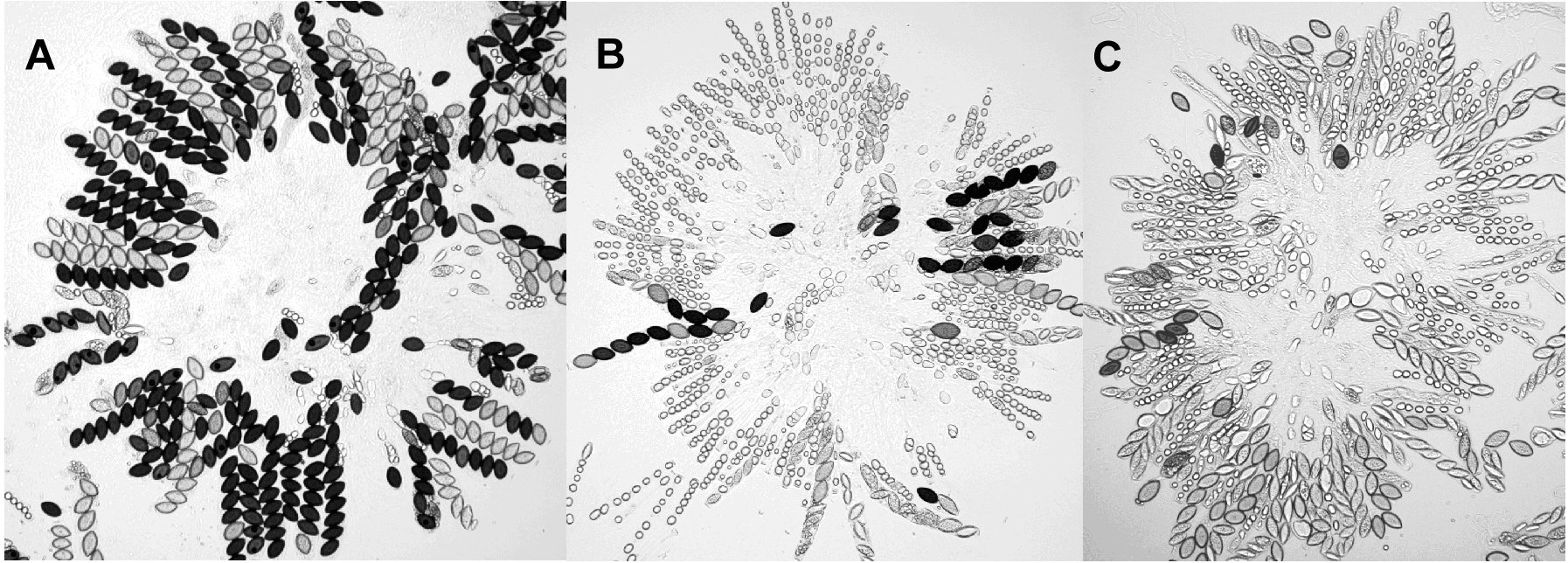
Figure 4. Examples of rosettes from control cross (A), otherwise isogenic Dmsh-4 heterozygote (B) and Dmsh-4 homozygote (C).
In conclusion, deletion of the putative Neurospora msh-4 gene interferes with sporogenesis and possibly meiosis. The deletion appears dominant, suggesting that msh-4 is normally expressed during meiosis and subject to meiotic silencing of unpaired DNA (Shiu and Metzenberg, 2002). The msh-4 candidate we deleted contains the MutS and ATPase domains common to all MSH4 orthologs and shares considerable predicted amino acid identity with both yeast and mammalian MSH4 proteins. Since we have now demonstrated a role for this gene in the sexual phase it is likely that it is indeed the Neurospora ortholog of MSH4.
Acknowledgements
This work was supported by grants from the Australian Research Council and the Flinders University Research Budget.
Alani, E., S. Lee, M. F. Kane, J. Griffith and R. D. Kolodner, 1997. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J. Mol. Biol. 265: 289-301.
Borkovich, K. A., et al., 2004 Lessons from the Genome Sequence of Neurospora crassa: Tracing the Path from Genomic Blueprint to Multicellular Organism. Microbiol. & Mol. Biol. 68: 1-108.
Catlett, N. L., Bee-Na Lee, O. C. Yoder and B. Gillian Turgeon, 2003. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Newslett. 50: 9-11.
Haber, L. T. and G. C. Walker, 1991. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 10: 2707-2715.
Hollingsworth, N. M., L. Ponte and C. Halsey, 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9 :1728-1739.
Hunter, N. and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing over during meiosis. Genes Dev.11 :1573-1582.
Marchler-Bauer, A. and S. H. Bryant, 2004. CD-Search: protein domain annotations on the fly. Nucl. Acids Res. 32 : 327-331.
Neyton, S., F. Lespinasse, P. Moens, P. Gaudray, V. Paquis-Flucklinger and S. Santucci-Darmanin, 2004. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol. Human Reprod. 10: 917-924.
Novak, J. E., P. B. Ross-Macdonald and G. S Roeder, 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013-1025.
Perkins, D. D. and E. G. Barry, 1977. The cytogenetics of Neurospora. Adv. Genet. 19: 133-285
Pochart, P., D. Woltering and N. M. Hollingsworth, 1997. Conserved Properties between Functionally Distinct MutS Homologs in Yeast. J. Biol. Chem. 272: 30345-30349.
Ross-Macdonald, P. and G. S. Roeder, 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069-1080.
Shiu, P. K. T. and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161: 1483-1495.
Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153 :1271-1283.
Return to the FGN 53 Table of Contents