Sequences important for heterokaryon incompatibility function in MAT A-1 of Neurospora crassa
Patrick K.T. Shiu1 and N. Louise Glass2
1Division of Biological Sciences, University of Missouri, Columbia, MO 65211. 2Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720
To whom correspondence may be addressed: shiup@missouri.edu or lglass@nature.berkeley.edu
Fungal Genetics Newsletter 53: 15-19.
Using chimeric constructs between the Neurospora crassa mat A-1 gene and the Podospora anserina FMR1 gene, we identified the amino acids important for the heterokaryon incompatibility function in the mating-type protein MAT A-1.
Strains of Neurospora crassa exist as two alternative mating type forms, A and a; differences in mating type are required for the initiation of the sexual cycle (Shiu and Glass, 2000). The mating-type (mat) locus also acts as a heterokaryon incompatibility (het) locus, such that hyphal fusion between A and a strains results in a heterokaryon that shows extremely inhibited growth, absence of conidiation, and hyphal compartmentation and death (Glass et al., 2000). The A and a mating type sequences occupy the same locus in A and a strains, but are highly dissimilar in sequence. The mat a-1 gene, which encodes a putative HMG (high mobility group) type of transcriptional regulator, provides all the functions for the a mating type, including mating, ascospore formation, and heterokaryon incompatibility (Chang and Staben, 1994). The mat A locus encodes three proteins. MAT A-2 and MAT A-3 are responsible for ascospore formation (Ferreira et al., 1998); MAT A-3 is a putative HMG type of transcriptional regulator. MAT A-1 is predicted to be a α-domain type of transcriptional regulator and is both necessary and sufficient to confer A mating specificity and trigger heterokaryon incompatibility with a strains (Glass et al., 1990). Mutations in an unlinked locus, tol, suppress mating-type incompatibility such that tol A and tol a strains are capable of forming a vigorous heterokaryon (Newmeyer, 1970; Shiu and Glass, 1999).
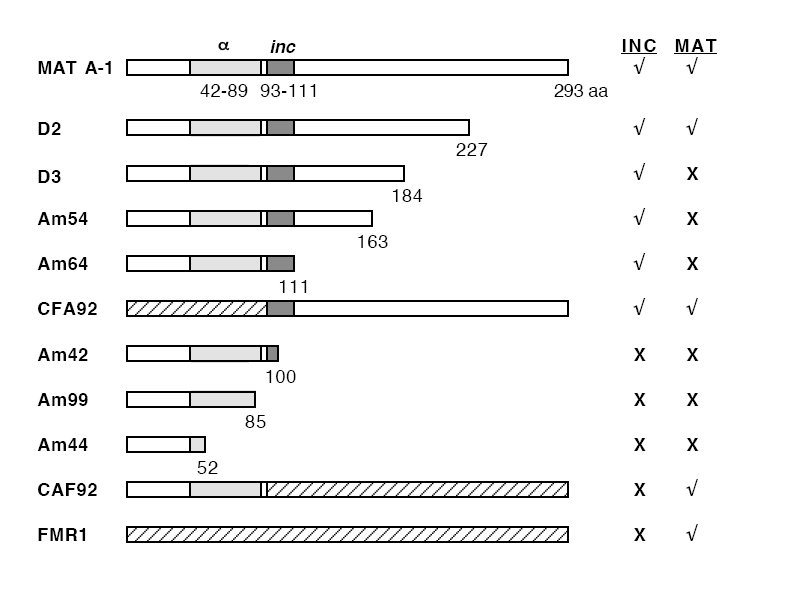
To determine the region required for the heterokaryon incompatibility function in MAT A-1 (293 amino acids), a series of nonsense, frameshift, and deletion constructs were evaluated for their ability to trigger heterokaryon incompatibility (Saupe et al., 1996). The results showed that a region from amino-acid position 1 to 111 is sufficient to confer the incompatibility function of MAT A-1. To further define the region important for the incompatibility function in MAT A-1, we expanded our previous studies with several additional constructs.
FMR1 does not mediate heterokaryon incompatibility in a transformation reduction assay:
The Podospora anserina FMR1 (fertilization minus regulator) gene confers mating identity for the mat- strain and is a homolog of N. crassa mat A-1 (Debuchy and Coppin, 1992). The two mating-type proteins are functional homologs since the introduction of P. anserina FMR1 confers mating in N. crassa and the introduction of N. crassa mat A-1 confers mating in P. anserina (Arnaise et al., 1993). In order to test if the FMR1 gene confers heterokaryon incompatibility in N. crassa in a transformation reduction assay, the FMR1 gene from plasmid pBPLP-0 (Debuchy et al., 1993) was cloned into pCB1004, a Neurospora-compatible vector containing a hygromycin-resistance marker (Carroll et al., 1996). The FMR1/pCB1004 plasmid was then subjected to the transformation reduction assay as described in Saupe et al (1996). Briefly, if a construct confers mat A-1 heterokaryon incompatibility activity, the transformation frequency should be significantly lower in the a recipient than in the A recipient, because transformants containing the incompatible mat a-1 and mat A-1 genes do not regenerate.
Transformation results show that the FMR1 construct has comparable transformation frequencies when introduced into both A and a spheroplasts (Figure 1). These results show that the FMR1 gene does not confer heterokaryon incompatibility (with mat a-1) in our transformation assay, although it confers mating with a N. crassa a strain (Arnaise et al., 1993). These data agree with previous results by Arnaise et al. (1993), which have shown that when FMR1 was introduced into N. crassa, an incompatibility reaction was not elicited.
Chimeric constructs between mat A-1 and FMR1:
Since FMR1 confers mating activity in Neurospora as mat A but does not confer heterokaryon incompatibility, it can be viewed as a mat A-1 mutant in the heterokaryon incompatibility function. However, although MAT A-1 and FMR1 are similar, there are many amino-acid differences between the two polypeptides, making it difficult to identify differences between the two polypeptides that correlate with incompatibility function in MAT A-1, but the lack of it in FMR1. The testing of chimeric proteins constructed between MAT A-1 and FMR1 is therefore the next logical approach to pinpoint the heterokaryon incompatibility domain.
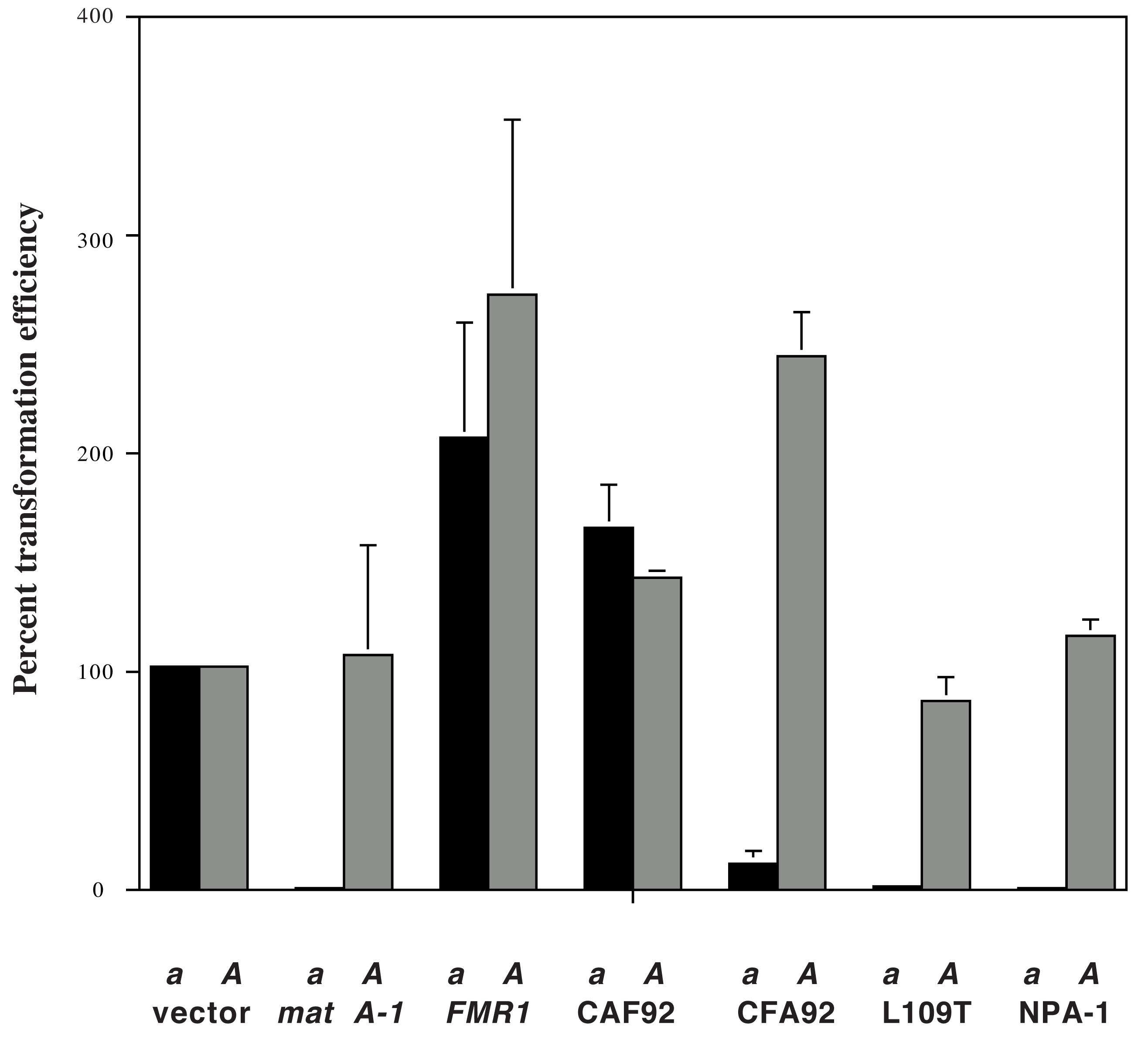
Figure 1. Results of the introduction of various constructs into a (pan-2; inl; arg-5 a) and A (pan-2; arg-5 A) recipient strains in the transformation reduction assay. CAF92 and CFA92 are chimeras constructed with the N. crassa mat A-1 and P. anserina FMR1 gene. CAF92 encodes amino acids 1-92 of MAT A-1 and 89-305 of FMR1. CFA92 encodes amino acids 1-88 of FMR1 and 93-293 of MAT A-1 (including the inc region of MAT A-1). L109T represents a site-directed mutant of mat A-1. NPA-1 represents the mat A-1 gene from N. pannonica. The transformation frequency for each transformation test is illustrated as a percentage of the transformation efficiency of the vector control. Transformation frequency of a hundred percent corresponds to the number of transformants that can be obtained when the vector control gives 100 transformants per 200 ng of DNA (introduced into 4 x 106 spheroplasts) in a transformation test. For the vector control, the typical number of transformants per plate (500 ng of DNA into 107 spheroplasts) is roughly 250. Results correspond to the mean of three transformations, with error bars indicating standard deviations.
From the results of the previous study, a construct containing the first 111 amino acids of MAT A-1 confers incompatibility function with a strains (Saupe et al., 1996; Figure 2). This segment contains the a-domain (position 42-89), which is found in many mating-type proteins (Shiu and Glass, 2000). We hypothesize that the region after the α-domain, which spans position 93-111 and is variable among different mating-type proteins, is important for the incompatibility function. To examine if this region (position 93-111), hereafter referred to as the inc region, is functionally important in mediating incompatibility, chimeras between MAT A-1 and FMR1 with a fusion point at the beginning of the inc region (i.e. between amino acids V92 and Y93) were constructed. Since there are no convenient restriction sites, an artificial Bst1107I site was introduced into both mat A-1 and FMR1 clones in order to facilitate chimeric construction. Two plasmids containing reciprocal chimeric genes, the CFA92 (pCB1004 vector containing amino acids 1-88 of FMR1 and 93-293 of MAT A-1) and CAF92 (pCB1004 vector containing amino acids 1-92 of MAT A-1 and 89-305 of FMR1), were constructed and used in the transformation assay.
When introduced into N. crassa competent cells, the CAF92 plasmid exhibited similar transformation efficiencies in A and a spheroplasts, whereas the CFA92 plasmid exhibited a considerably lower transformation frequency (20-fold) in the a recipient (Figure 1). These data indicate that the CFA92 construct (containing the MAT A-1 inc region) confers heterokaryon incompatibility in N. crassa whereas CAF92 does not. Furthermore, when CFA92 and CAF92 were introduced into a tol a recipient (ad-3A nic-2; tol a), CFA92- and CAF92- transformants conferring dual mating activity were recovered (i.e. they initiated perithecial development with both the A and a tester strains). These data show that functions of mating and incompatibility in MAT A-1 can be separated. The absence of incompatibility function in CAF92 is due to lack of an incompatibility domain (the MAT A-1 inc region) and not due to loss of function of MAT A-1. The position of the breakpoint in the two chimeric constructs and their transformation/mating results show that the differences in the inc region were responsible for the lack of incompatibility function in the FMR1 gene. A summary of our previous and present results can be found in Figure 2.

Figure 2. mat A-1 constructs and their functional analyses. Assayed functions include mating-type incompatibility (INC) and male mating (MAT) activity. Detailed topography of mutations and translational products of the Am mutants as well as the deletion constructs (D2 and D3) can be found in Saupe et al (1996). CFA92 and CAF92 are chimeric proteins constructed from mat A-1 and FMR1. The a-domain is a region of homology found in many mating-type proteins. The incompatibility activity is correlated with the inc region (position 93-111) in MAT A-1.
Site-directed mutagenesis of the inc region in mat A-1:
Our previous results show that the difference in the inc region between MAT A-1 and FMR1 were responsible for the lack of the incompatibility function in FMR1. One potential candidate is the L109T substitution (Figure 3). L109 is a conserved leucine among different Neurospora MAT A-1 homologs in the inc region. This leucine residue at position 109 is replaced by a threonine in the FMR1 polypeptide. To test if the L109T substitution is responsible for the loss of heterokaryon incompatibility in FMR1, the same amino-acid substitution was introduced to the wild-type mat A-1 gene. The site-directed mutant was subjected to the transformation assay. Results of the assay indicate that the L109T mutant confers heterokaryon incompatibility (Figure 1). Therefore, the L109T substitution is not solely responsible for the absence of the incompatibility function in FMR1.
93 111
| |
N. crassa YSSIRTYLEQEKVTLQLWI
N. terricola YSSIRTYLEQEKVTLQLWI
N. africana YSSIRTYLEQEKVTLQLWI
N. pannonica YSAIRTYLEEEKVNLQLWN
P. anserina YSAIRDQLAEQNVTLQTWI
Figure 3. Amino-acid position 93-111 in Neurospora crassa MAT A-1 and its homologs in other fungal species. The numbers indicate the amino-acid position according to the N. crassa MAT A-1 protein. Podospora anserina FMR1 is the only protein listed here that does not confer heterokaryon incompatibility function. The a-domain of MAT A-1 is found from position 42 to 89 (not shown), 4 amino acids upstream of the above region. The bold amino acids are different between MAT A-1 and FMR-1. The underlined amino acids are shown to be not solely responsible for the lack of incompatibility function in FMR1.
Heterokaryon incompatibility function in Neurospora pannonica mat A-1:
The L109T site-directed mutagenesis did not reveal a disruption of incompatibility function in MAT A-1. However, there are other candidates within the inc region that could explain the “compatible” phenotype of FMR1. As shown in Figure 3, there are many other changes between FMR1 and the Neurospora MAT A-1 within the inc region, namely S95A, T98D, Y99Q, E101A, Q102E, E103Q, and K104N. One of the residues mentioned above is of special interest: S95 is the only potential protein kinase phosphorylation site (S/T-X-R/K) in the MAT A-1 protein. It is possible that the S95A change in FMR1 leads to the absence of phosphorylation at that position and therefore prevents FMR1 to mediate incompatibility.
It is possible to test the importance of the S95A substitution without the use of site-directed mutagenesis, since the Neurospora pannonica mat A-1 gene contains the same S95A substitution (Vellani, 1998). The N. pannonica mat A-1 gene from FGSC (McCluskey, 2003) strain 7221 was amplified, cloned and subjected to the transformation assay as described above. Our results indicate that the N. pannonica mat A-1 gene confers heterokaryon incompatibility in N. crassa (Figure 1, NPA-1). Therefore the S95A substitution is not solely responsible for the loss of incompatibility function in the FMR1 polypeptide. Because the N. pannonica mat A-1 gene also contains the Q102E substitution (as found in FMR1), the same conclusion can also be drawn for that amino acid difference.
In conclusion, our present study provides evidence that a region from position 93 to 111 is important for the heterokaryon incompatibility function in MAT A-1. Using chimeric constructs between mat A-1 and FMR-1, we have identified several amino acids that could be important for this function. Our subsequent studies show that five amino-acid differences between MAT A-1 and FMR1 (T98D, Y99Q, E101A, E103Q, and K104N) could explain the lack of incompatibility function in FMR1.
Acknowledgements
We are thankful for the support from the Natural Sciences and Engineering Research Council (to NLG) and from the University of Missouri Research Board (to PKTS). We are grateful to the FGSC for the pCB1004 plasmid and to Robert Debuchy for the pBPLP-0 plasmid.
References
Arnaise, S., D. Zickler, and N.L. Glass. 1993. Heterologus expression of mating-type genes in filamentous fungi. Proc. Natl. Acas. Sci. USA 90: 6616-6620.
Carroll, A.M., J.A. Sweigard, and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 41: 22.
Chang, S. and C. Staben. 1994. Directed replacement of mt A by mt a-1 effects a mating type switch in Neurospora crassa. Genetics 138: 75-81.
Debuchy, R. and E. Coppin. 1992. The mating types of Podospora anserina: functional analysis and sequence of the fertilization domains. Mol. Gen. Genet. 233: 113-121.
Debuchy, R., S. Arnaise, and G. Lecellier. 1993. The mat- allele of Podospora anserina contains three regulatory genes required for the development of fertilized female organs. Mol. Gen. Genet. 241: 667-673.
Ferreira, A.V.B., Z. An, R.L. Metzenberg, and N.L. Glass. 1998. Characterization of mat A-2, mat A-3 and DmatA mating-type mutants of Neurospora crassa. Genetics 148: 1069-1079.
Glass, N.L., J. Grotelueschen, and R.L. Metzenberg. 1990. Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. USA 87: 4912-4916.
Glass, N.L., D.J. Jacobson, and P.K.T. Shiu. 2000. The genetics of hyphal fusion and post fusion recognition in filamentous ascomycete fungi. Annu. Rev. Genet. 34: 165-186.
McCluskey, K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52: 245-262.
Newmeyer, D. 1970. A suppressor of the heterokaryon-incompatibility associated with mating type in Neurospora crassa. Can. J. Genet. Cytol. 12: 914-926.
Saupe, S., L. Stenberg, K.T. Shiu, A.J.F. Griffiths, and N.L. Glass. 1996. The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol. Gen. Genet. 250: 115-122.
Shiu, P.K.T. and N.L. Glass. 1999. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics 151: 545-555.
Shiu, P.K.T. and N.L. Glass. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Micro. 3: 183-188.
Vellani, T.S. 1998. Positional regulation and evolution of mating type genes in heterothallic and homothallic species of Neurospora. Ph.D. Thesis, University of British Columbia, Vancouver, Canada.
Return to the FGN 53 Table of Contents