Podospora anserina AS6 gene encodes the cytosolic ribosomal protein of the E. coli S12 family
Michelle Dequard-Chablat1 and Philippe Silar1, 2
1Institut de Génétique et Microbiologie, Bât. 400 Université de Paris 11,
91405 Orsay cedex, France.
2UFR de Biochimie, Université de Paris 7 - Denis Diderot, case 7006, 2
place Jussieu, 75005, Paris, France
To whom correspondence may be addressed: michelle.chablat@igmors.u-psud.fr or
philippe.silar@igmors.u-psud.fr
Fungal Genetics Newsletter 53: 26-29
The ribosomal proteins of the E. coli S4, S5 and S12 families that are part of the ribosome accuracy center control translation accuracy both in prokaryotes and eukaryotes. In Podospora anserina, genes coding for S4 and S5 have already been identified. Here, we identify the gene coding for the S12 protein homologue and show that it is identical to the genetically known AS6 gene.
Podospora anserina has been used in intensive search of translation accuracy mutants (Coppin-Raynal et al. 1988). Several factors involved in the maintenance of accuracy have been identified in this organism including the tRNA suppressors su4-1 and su8-1 (Debuchy et al. 1985), as well as elongation factor eEF1A coded by AS4 (Silar et al. 1994), termination factors eRF1 and eRF3 coded by su1 and su2/AS2 respectively (Gagny et al. 1998), ribosomal proteins S12 coded by AS1 (Dequard-Chablat et al. 1994), S7 coded by su12 (Silar et al. 1997) and S1 coded by su3 (Silar et al. 2003). S12, S7 and S1 refer to the P. anserina numbering for ribosomal proteins (Dequard-Chablat et al. 1986) since the su12 and su3 genes code for the ribosomal proteins homologues of the E. coli S4 and S5, respectively. These two proteins are part of an accuracy center that has been conserved for more than two billion years in both prokaryotes and eukaryotes (Alksne et al. 1993). The center contains a third protein corresponding to the E. coli S12 protein, which remains to be identified in P. anserina. This protein is highly conserved and essential in all prokaryotes and eukaryotes investigated to date (Alksne et al. 1993).
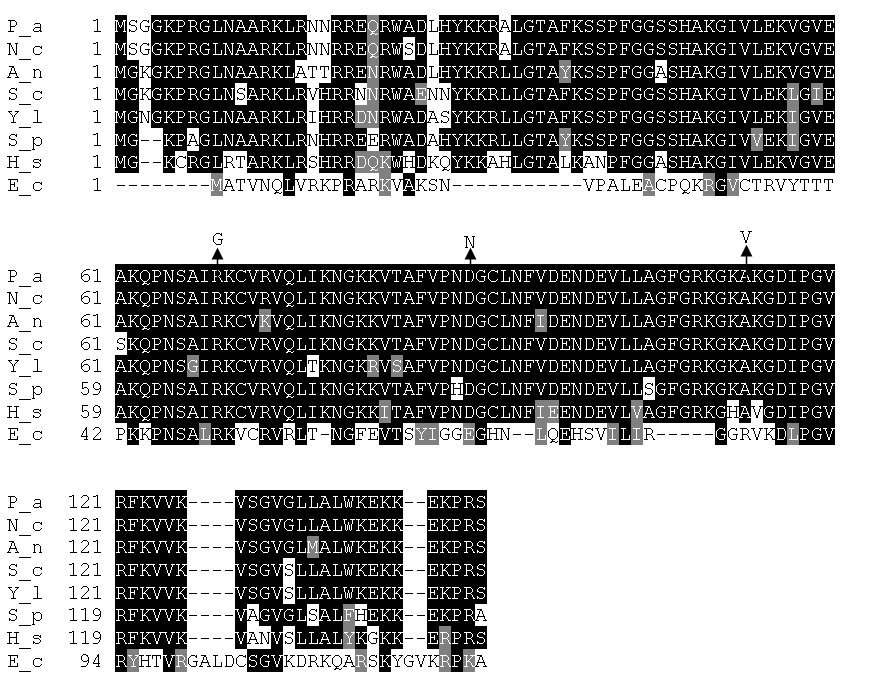
To identify the gene coding this protein in P. anserina, we took advantage of the availability of the complete genomic sequence of this fungus (available at http://podospora.igmors.u-psud.fr). We first located on the sequence map the gene encoding the P. anserina protein of the S12 family that would act in cytosolic translation. To do this, we searched the genome sequence by BLAST using the Saccharomyces cerevisiae S28 protein, which is a eukaryotic homolog of the E. coli S12 protein (Alksne et al. 1993). A single CDS, Pa_3_536, was obtained with a significant score (10-72; Figure 1). It has 89% identity and 94% similarity with the S. cerevisiae S28 protein. At least one orthologue is present in the complete genomes of all organisms sequenced to date and the gene is highly conserved in fungi (figure 1). For example, the P. anserina and Neurospora crassa proteins are 99% identical .
Pa_3_5360 is located on the right arm of chromosome 3 in a region that would segregate with a 70% second division frequency, strongly suggesting that it could correspond to the CDS of the previously known AS6 gene since (1) AS6 is located at this position on the genetic map (Picard-Bennoun et al. 1980), (2) AS6 controls translation accuracy (Picard-Bennoun 1981), (3) numerous mutations of this gene were recovered as suppressors of su12 mutations and thus (Dequard-Chablat 1986) and (4) AS6 encodes a ribosomal protein (named S19 in the P. anserina nomenclature; Dequard-Chablat et al. 1986).
To confirm this, we first sequenced the three alleles of Pa_3_5360 present in the AS6-1, AS6-2 and AS6-5 mutants. DNA fragments were amplified using oligonucleotides 5'- ttgcgagatcccatcaaag-3' and 5'-agcctcgcagactcctca-3' and the three mutant genomic DNAs as template. Complete sequence of the DNA fragments was then performed and compared with that of wild type. AS6-1 contained a C to G transversion that changes arginine 73 to glycine, AS6-2 a G to A transition that changes aspartate 90 to asparagine and AS6-5 a C to T transition that changes alanine 113 to valine (Figure 1). Finally, we co-transformed the AS6-1 mutant with plasmid pAS6 (plasmid GA0AA399BE01 from the P. anserina genome sequence project) containing a DNA fragment carrying the AS6+ allele and the pBC-hygro vector carrying a hygromycin B resistance gene (Silar 1995). AS6-1 has a strong sexual defect since it is unable to differentiate perithecia (Figure 2). No transformants with a restored wild-type phenotype was recovered among the 30 retrieved in the control experiment with the pBC-hygro vector alone, whereas, six transformants with a restored wild-type phenotype were recovered among 13 analyzed in the transformation with both pBC-hygro and pAS6 (Figure 2) further demonstrating that Pa_3_5360 is the CDS corresponding to AS6.

Figure 1. Comparison of the P. anserina S12 homologue (P_a) with orthologues in Neurospora crassa (N_c), Aspergillus nidulans (A_n), Saccharomyces cerevisiae (S_c), Yarrowia lipolytica (Y_l), Schizosaccharomyces pombe (S_p), Homo sapiens (H_s) and Escherichia coli (E_c). Arrows point the positions of the AS6 mutations in the P. anserina protein.
Figure 2. Perithecium production in the indicated strains.
In conclusion, the AS6 gene codes the ribosomal protein of the E. coli S12 family acting in cytosolic translation. In P. anserina, mutations of this gene lead to an increased in translation accuracy (Picard-Bennoun 1981; Dequard-Chablat 1986; Dequard-Chablat et al. 1986), which in turn has many physiological effects. Indeed, it was demonstrated that increase translation accuracy in the AS6-5 mutant triggers a specific suppressive effect on the accumulation of the circular senDNAá in the mitochondria during P. anserina senescence (Silar et al. 1997), entails a strong defect in sexual reproduction (Coppin-Raynal et al. 1988; Dequard et al. 1984) and permits the development of the epigenetic cell degeneration Crippled Growth (Haedens et al. 2005; Kicka et al. 2004; Silar et al. 1999). In all these cases, it was demonstrated that increase accuracy was responsible for the defects, indicating the translation errors are necessary for normal P. anserina physiology as postulated previously (Picard-Bennoun, 1982).
Acknowledgment.
University of Paris 11 and C.N.R.S. intramural funds supported this work.
References
Alksne, L.E., R.A. Anthony, S.W. Liebman and J.R. Warner, 1993. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci U S A. 90, 9538-41.
Coppin-Raynal, E., M. Dequard-Chablat and M.,Picard, 1988, Genetics of ribosomes and translational accuracy in Podospora anserina, in:M. Tuite, M. Picard,M. Bolotin-Fukuhara, Genetics of translation: new approaches,Springer-Verlag,pp. 431-442.
Debuchy, R. and Y. Brygoo, 1985. Cloning of opal suppressor tRNA genes of a filamentous fungus reveals two tRNASerUGA genes with unexpected structural differences. EMBO J. 4, 3553-6.
Dequard, M. and E. Coppin, 1984. Increase of translational fidelity blocks sporulation in the fungus Podospora anserina. Mol. Gen. Genet. 195, 294-300.
Dequard-Chablat, M., 1986. Genetics of translational fidelity in Podospora anserina: are all the genes involved in this ribosomal function identified? Curr Genet. 10, 531-6.
Dequard-Chablat, M., E. Coppin-Raynal, M. Picard-Bennoun and J.J. Madjar, 1986. At least seven ribosomal proteins are involved in the control of translational accuracy in a eukaryotic organism. J Mol Biol. 190, 167-75.
Dequard-Chablat, M and C.H. Sellem, 1994. The S12 ribosomal protein of Podospora anserina belongs to the S19 bacterial family and controls the mitochondrial genome integrity through cytoplasmic translation. J Biol Chem. 269, 14951-6.
Gagny, B. and P. Silar, 1998. Identification of the genes encoding the cytosolic translation release factors from Podospora anserina and analysis of their role during the life cycle. Genetics. 149, 1763-75.
Haedens, V., F. Malagnac and Silar, 2005. Genetic control of an epigenetic cell degeneration syndrome in Podospora anserina. Fungal Genet Biol. 42, 564-77.
Kicka, S. and P. Silar, 2004. PaASK1, a Mitogen-Activated Protein Kinase Kinase Kinase That Controls Cell Degeneration and Cell Differentiation in Podospora anserina. Genetics. 166, 1241-1252.
Picard-Bennoun, M., 1981. Mutations affecting translational fidelity in the eukaryote Podospora anserina: characterization of two ribosomal restrictive mutations. Mol Gen Genet. 183, 175-80.
Picard-Bennoun, M., 1982. Does translational ambiguity increase during cell differentiation ? FEBS Letters. 149, 167-170.
Picard-Bennoun, M. and D. Le Coze, 1980. Search for ribosomal mutants in Podospora anserina: genetic analysis of cold-sensitive mutants. Genet Res. 36, 289-97.
Silar, P., 1995. Two new easy-to-use vectors for transformations. Fung. Genet. Newslett. 42, 73.
Silar, P., C. Barreau, R. Debuchy, S. Kicka, B. Turcq, A. Sainsard-Chanet, C.H. Sellem, A. Billault, L. Cattolico, S. Duprat and J. Weissenbach, 2003. Characterization of the genomic organization of the region bordering the centromere of chromosome V of Podospora anserina by direct sequencing. Fungal Genet Biol. 39, 250-63.
Silar, P., V. Haedens, M. Rossignol and H. Lalucque, 1999. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics. 151, 87-95.
Silar, P., F. Koll, M. Rossignol, 1997. Cytosolic ribosomal mutations that abolish accumulation of circular intron in the mitochondria without preventing senescence of Podospora anserina. Genetics. 145, 697-705.
Silar, P. and M. Picard, 1994. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J Mol Biol. 235, 231-6.
Return to the FGN 53 Table of Contents
Return to the FGSC main page