
Detection of physical interactions by immunoprecipitation of FLAG- and HA-tagged proteins expressed at the his-3 locus in Neurospora crassa
Tsuyoshi Kawabata and Hirokazu Inoue
Laboratory of Genetics, Department of Regulation Biology
Saitama University, Saitama City Japan
Fungal Genetics Newsletter 54:5-8
Protein function is often regulated through interactions with other protein(s) or by post-translational modifications. To understand these mechanisms, it is useful to utilize antibodies. However, it is not always certain whether a good antibody can be made for this purpose. The use of epitope tags eliminates the troubles associated with raising antibodies. In this report, we present a method to detect interactions between proteins by using two types of epitope-tagged proteins, FLAG- and HA-tagged proteins in Neurospora. These constructs were introduced at and expressed from the his-3 locus in different strains. To examine protein-protein interactions, heterokaryons between these strains were constructed. We conclude that this strategy is a useful tool to investigate protein function and protein interactions.
After publication of a draft of the Neurospora genome project (Galagan et al., 2003), gene knockout (Colot et al., 2006) and DNA array projects (Kasuga et al., 2005) started to elucidate gene function. Information and materials obtained from these projects have substantially changed experimental methods and accelerated molecular studies in Neurospora. Studies of protein-protein interaction and post-translational modifications such as ubiquitylation, SUMOylation, phosphorylation and methylation are indispensable. In many cases, one needs to use specific antibodies to examine protein interactions and modifications. The experiments that determine protein-protein interactions by using epitope tags therefore have been carried out in Neurospora (Dementhon et al., 2006; Sarkar et al., 2002).
In this report, we present useful targeting vectors to express FLAG- or HA-tagged proteins of interest integrated at the his-3 locus in N. crassa. To create the vectors, we amplified a DNA fragment that contained sequences of linker and FLAG-epitope tag by PCR, then digested it with SpeI and BamHIand inserted the DNA fragment into the SpeI/BamHI site of the his-3 targeting vector pMF272, (Freitag et al., 2004), resulting in plasmid pFLAGN1. Similarly, a DNA fragment encoding HA-epitope tag was digested with SpeI and XbaI and ligated into the SpeI/XbaIsite of pMF272, resulting in the creation of pHAN1. Confirmation that the sequence of interest was inserted into the vector was performed by DNA sequencing.

Figure 1. Construction of the vectors. DNA sequence containing linker and epitope tag was inserted between ccg-1 promoter and sgfp+-ORF of pMF272. The plasmids were constructed to add the epitope tags to a polypeptide at its N-terminus, not its C-terminus.
We selected PCNA as a model for detecting a physical interaction between proteins. In S. cerevisiae, PCNA is an essential component for DNA replication (Johnson and O'Donnell, 2005) and also functions in DNA repair (Hoege et al., 2002) and gene silencing (Zhang et al., 2000). PCNA exists in the cell as a homotrimeric ring, which functions as a sliding clamp and a docking station for many proteins. Formation of the trimer is essential for the functions (Jonsson et al., 1995). Neurospora PCNA is also essential for its vegetative growth, as indicated from the result of gene-targeting experiment (data not shown). To determine Neurospora PCNA interacts with each other in vivo, cDNA of Neurospora PCNA was cloned, digested with BamHI/EcoRIand ligated into BamHI/EcoRI-digested pFLAGN1 or pHAN1 to generate pFGNPCNA or pHANPCNA, respectively. Each of them were linearized by a digestion with NdeI and transformed into the his-3 inl strain (FGSC#10376, KBT-H3-26A; mat A ridΔ::hph+ his-3::lplΔ(5192-6046) ::hph+::tk+; inl) or his-3 pan-2 strain (FGSC#10375, KBT-H3-4A; mat A ridΔ::hph+ his-3::lplΔ(5192-6046) ::hph+::tk+ al-2; pan-2; cot-1) by electroporation as described previously (Margolin et al., 1997). The his-3 inl strain was derived from a cross of the his-3 mutant strain (FGSC#9097; mat A his-3::lplΔ(5192-6046) ::hph+::tk+; inl) (Lee et al., 2003) with a rid-defective strain (mat a ridΔ::hph+) and the his-3 pan-2 strain was derived from a cross of the his-3 inl strain with 74-OR31-14a (FGSC# 4934; mat a al-2; pan-2; cot-1). Resulting histidine-prototroph transformants were isolated and grown for 7 days at 30∘C.
To make the transformant homokaryotic, single colony isolation was repeated several times. The genomic DNA was isolated to determine whether the expression construct was integrated correctly at the his-3 locus and the isolated clones were homokaryon by PCR. The clones were named KBT-FGPC-11A (FGSC#10377; mat A ridD::hph+ his-3::lplΔ(5192-6046) ::hph+::tk+::flag-pcna+; inl) and KBT-HAPC-7A (FGSC#10374; mat A al-2 ridΔ::hph+; his-3::lplΔ(5192-6046) ::hph+::tk+::ha-pcna+; pan-2; cot-1), respectively. The his-3 (FGSC#9097) strain was generated to allow for rapid identification of homokaryotic transformants containing the expected gene replacement event (Lee et al., 2003). Vectors used in this study suit for targeting event in both the his-3 mutant strain (FGSC#9097) and a canonical his-3 strain (such as FGSC#6103), but not adjusted to gain the advantage of the his-3 mutant strain (FGSC#9097) due to a lack of suitable homology sequence, resulting in remaining of hph+::tk+ sequence in its genome.

Figure 2. Procedure for investigating interaction between HA-tagged protein and FLAG-tagged protein expressed at the his-3 locus. Western blotting was performed with cell extract from mycelia of each strain. Heterokaryotic strain was generated by a culture of mixed strains, the KBT-FGPC-11A and KBT-HAPC-7A, in the same test tube.
In order to analyze the interaction between FLAG-PCNA and HA-PCNA, two homokaryon strains, KBT-FGPC-11A and KBT-HAPC-7A, and the heterokaryon strain were analyzed (Fig. 2). Conidia of each strain were incubated in 50 ml of liquid medium (final 2.0×106 conidia/ml) containing nutrients as indicated in Figure 2 and cultured with shaking at 30∘C for 12 hours. Cells were collected and homogenized with Micro SmashTM MS-100 (Tomy) in a buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 10% glycerol, 0.3% TritonX100) containing a protease inhibitor (Complete, EDTA-free: Roche). The extract was centrifuged at 14000 rpm for 10 min at 4∘C. The supernatant was mixed with anti-FLAG M2 Affinity Gel (Sigma) for 1 hour at 4∘C. The precipitates and whole cell extract were electrophoresed in 11% SDS-PAGE gels. Following transfer from the gels onto PVDF membranes, they were probed with mouse anti-FLAG M2 monoclonal antibody (Sigma) or anti-HA monoclonal antibody (HA.11: Covance).
FLAG-PCNA or HA-PCNA was detected in whole cell extracts from the KBT-FGPC-11A or KBT-HAPC-7A strains by anti-FLAG or -HA antibody, respectively, indicating that tagged PCNA was expressed in N. crassa (Fig. 3, left three lanes). FLAG-PCNA was detected with anti-FLAG antibody in the immunoprecipitated samples from both the heterokaryon and the FLAG-PCNA-expressing strain but not detected in that of HA-PCNA-expressing strain. HA-PCNA was detected with anti-HA antibody in the immunoprecipitated sample of the heterokaryon strain but was not detected in the immunoprecipitated samples from the strains expressing HA-PCNA or FLAG-PCNA only (Fig.3, right three lanes). This indicates that FLAG-PCNA and HA-PCNA physically interact with each other in Neurospora, and the interaction is not affected by HA-tag or FLAG-tag at its N-terminus.
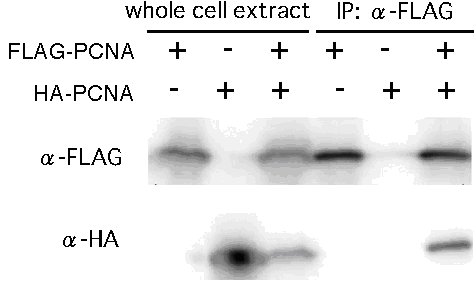
Figure 3. Examination of interaction between FLAG-PCNA and HA-PCNA by western blotting. The left threes lanes show Western blotting performed against whole cell extract and the right three lanes show that of IP-samples produced by using anti-FLAG antibody. In the right three lanes, HA-PCNA was detected only in the sample from the mixed strain that expressed both FLAG-PCNA and HA-PCNA.
In conclusion, it was shown that FLAG- or HA-tagged proteins expressed from the his-3 locus can be detected by western blotting and that physical interactions between proteins can be accessed by using a forced-heterokaryon strain. This is a useful method to analyze post-translational modification and physical interactions between proteins. The strains and vectors used in this study are available in FGSC.
Acknowledgements
We thank Yasushi Saeki and Akio Toh-e for help with western blotting. This research was supported by Rational Evolutionary Design of Advanced Biomolecules, Saitama Prefecture Collaboration of Regional Entities for Advancement of Technological Excellence, Japan Science and Technology Agency.
References
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., Weiss, R. L., Borkovich, K. A., and J. C. Dunlap, 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103, 10352-10357.
Domenthon, K., Iyer, G., and N. L. Glass, 2006. VIB is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryotic Cell 5, 2161-2173.
Freitag, M., Hickey, P. C., Raju, N. B., Selker, E. U., and N. D. Read, 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol 41, 897-910.
Galagan, J. E., et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859-868.
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135-141.
Johnson, A., and M. O'Donnell, 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem 74, 283-315.
Kasuga, T.,Townsend, J. P., Tian, C. Gilbert, L. B. Mannhaupt, G. Taylor, J. W., Glass, N. L. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination NAR 33, 6469-85
Jonsson, Z. O., Podust, V. N., Podust, L. M., and U. Hubscher, 1995. Tyrosine 114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J 14, 5745-5751.
Lee, D. W., Haag, J. R., and R. Aramayo, 2003. Construction of strains for rapid homokaryon purification after integration of constructs at the histidine-3 ( his-3) locus of Neurospora crassa. Curr Genet 43, 17-23.
Margolin, B. S., Freitag, M., and E. U. Selker, 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newslett 44, 34-36.
Sarkar, S., G. Iyer, J. Wu, and N. L. Glass. 2002. Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility in Neurospora crassa. EMBO J. 18:4841-4850
Zhang, Z., Shibahara, K., and B. Stillman, 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221-225.
Return to the FGN 54 Table of Contents