Two yeast plasmids that confer nourseothricin-dihydrogen sulfate and hygromycin B resistance in Neurospora crassa and Cryphonectria parasitica.
Robert Phillip Smith and Myron L. Smith
Nesbitt Biology Building, Department of Biology, Carleton University, Ottawa, Ontario, Canada.
Fungal Genetics Newsletter 54:12-13
Two plasmids that were previously used with yeast, pRS41N and pRS41H, were found to confer clonNAT and hygromycin B resistance, respectively, in the filamentous fungi Neurospora crassa and Cryphonectria parasitica. These plasmids are suitable for routine cloning and for use in forcing heterokaryons and are available through the FGSC.
Identification of novel resistance markers is important to further our ability to manipulate and characterize genetic elements in filamentous fungi. Hygromycin B (hygB) inhibits protein translation and has been used extensively as a selectable marker in filamentous fungi (Rao et al,, 1985). Alternative, affordable selectable markers are desirable for various applications, including for forcing heterokaryons. Previously, nourseothricin-dihydrogen sulfate (clonNAT) has been used as a selectable marker in fungi (Kück and Hoff, 2006). This aminoglycoside binds to ribosomes and results in inhibition and errors in protein synthesis (Cundliffe, 1989). Here we report that two vectors, pRS41N (clonNAT resistance) and pRS41H (hygB resistance), that were previously characterized in S. cerevisiae but not in filamentous fungi, are appropriate transformation vectors and suitable for forcing heterokaryons in Neurospora crassa and Cryphonectria parasitica.
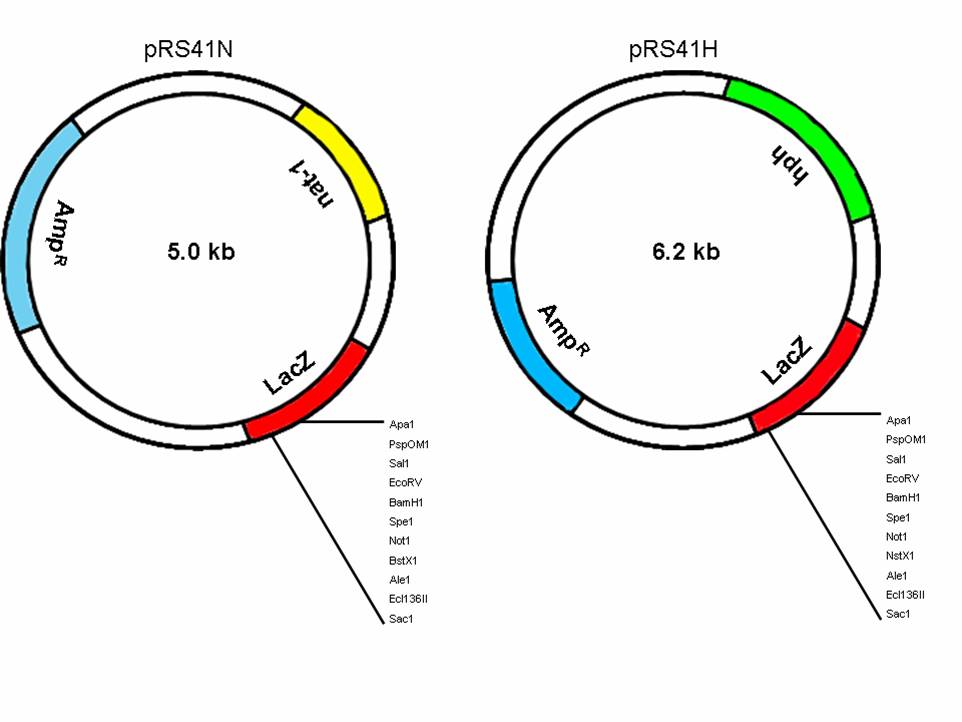
Figure 1. Yeast plasmids pRS41N and pRS41H that confer clonNAT and hygB resistance, respectively, to N. crassa and C. parasitica. Unique restriction sites in the multiple cloning site are indicated. (large version)
Plasmids pRS41N and pRS41H (Figure 1) were derived from pRS416 as described in Taxis and Knop (2006). Resistance genes in both plasmids are driven by the TEF promoter from Ashbya gossypii, and S. cerevisiae CYC1 and ADH1 terminators are used in pRS41H and pRS41N, respectively. This contrasts to previously characterized vectors such as pCB1004 (Carroll et al., 1994) and pD-NAT1 (Kuck and Hoff, 2006) that use Aspergillus nidulans trpC promoter and terminator. Transformations with pRS41N and pRS41H were done with a PEG-mediated transformation protocol (Smith et al., 2000). Selection of N. crassa, pRS41N transformants was on Vogel’s minimal medium containing 200 ug/ml of clonNAT (Werner Bioagents, Jena, Germany) and yielded ~20 colonies / ug plasmid DNA. Similarly, C. parasitica transformed with pRS41N, was selected on Potato Dextrose Agar (PDA) containing 100 ug/ml of clonNAT and ~10 colonies/ug of plasmid were recovered. Stable transformation frequencies of ~12 colonies/ug were achieved with both species using pRS41H by selection for resistance to hygB at 200 ug/ml and 30 ug/ml for N. crassa and C. parasitica, respectively.
Eleven unique restriction sites are available in the multi-cloning sites of pRS41N and pRS41H to facilitate blue-white selection. We used the SacI and SpeI restriction sites to clone un-24PA into pRS41N and transformed this construct into N. crassa strain C9-2, which contains un-24OR. The transformants had the characteristic self-incompatible phenotype (Micali and Smith, 2006) indicating that un-24PA was expressed as a transgene using this system. We further tested whether pRS41N and pRS41H could be used to force heterokaryons. Strains of both species were transformed separately with pRS41N and pRS41H. Small (~0.3 mm3) agar blocks containing hygB- and clonNAT-resistant hyphae were grown for two days on nonselective medium and then transferred to medium containing both clonNAT and hygB at the concentrations given above. Heterokaryon outgrowths of both species were evident within ~2 days and subsequently grew at near wild-type rates in clonNAT-hygB medium. We conclude that pRS41N and pRS41H are suitable as general transformation vectors and are particularly useful for generating forced heterokaryons in both fungal species.
Acknowledgements
This work was supported by OGS and NSERC CGS to RPS and an NSERC Discovery Grant to MLS. Thank you to Dr. Christof Taxis for the plasmids used in this study. Thanks to Dr. John Vierula for his helpful suggestions with this manuscript.
References
Carroll, A.M. Sweigard, J.A., and Valent, B. 1994 Improved vectors for selecting resistance to hygromycin. Fungal Genetics Newsletter. 41:22
Cundliffe, E. 1989. How antibiotic producing organisms avoid suicide. Annual Review of Microbiology. 43:207-231
Kück, U., and Hoff, B. 2006. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genetics Newsletter. 53:9-11.
Rao, R.N., Allen, N.E., Hobbs Jr., J.N., Alborn, W.E., Kirst, H.A., and Pashcal, J.W. 1983. Genetic and enzymatic basis of hygromycin B resistance in Escherichia coli. Antimicrobial Agents and Chemotherapy. 24: 689-695.
Micali, C.O., and Smith, M.L. 2006. A nonself recognition gene complex in Neurospora crassa. Genetics. 173: 1991-2004.
Smith, M.L., Micali, O.C., Hubbard, S., Mir-Rashed, N., Jacobson, D.J. and Glass, N.L. 2000. Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics. 155:1095-1104
Taxis, C., and Knop, M. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques. 40: 73-78
Return to the FGN 54 Table of Contents