Neurospora mutants beginning with P
You may jump directly to the following sections:
Peach (pe)
Phenylalanine (phe)
Permease, basic amino acid (pmb)
Proline (pro)
Pyrimidines (pyr)
Quinic acid (qa)
Recombination (rec)
Ropy (ro)
pa: pale
IR. Between cr-1 (10%) and dir (37%) (609, 610).
Conidia sparse, clumped, and pale. Photograph (610). (Stock lost. Possibly
wc-2?)
pab-1: p-aminobenzoic acid-1
VR. Between inl (1 to 10%) and met-3 (1 to 2%) (3629 1036).
(47)
Requires p-aminobenzoic acid (1057) (Fig. 11). Apparently
cannot use folate
(mono- or triglutamate) (1179).
pab-2: p-aminobenzoic acid-2
VR. Right of ad-7 (8%). Left of inv (3%) and asn (1 to
15%). Linked to ro-4
(0/407) (156, 816, 818, 918, 1036). (47)
Requires p-aminobenzoic acid (1182) (Fig. 11). Allele
71301 called pab-3
(1182); shown to be allelic by Drake (289).
pab-3
See pab-2.
pan-1: pantothenic acid-1
IVR. Between ad-6 (1 to 2%) and cot-1 (2 to 3%) (633, 692, PB).
(482). cel,
col-1, int, pho-3, and thi-5 all appear to be closely linked in this crowded
region.
Requires intact pantothenic acid for growth under standard conditions. Able
to synthesize both precursors, beta-alanine and pantoyl lactone (1058). Ability
to synthesize pantothenic acid from beta-alanine plus pantoyl lactone is
demonstrable in vitro but not in vivo unless cultures are aerated (1111, 1113,
1114). Unlike pan-2, pan-1 has no effect on ascospore ripening in
heterozygous crosses. Called group A. For alleles see reference 138.
pan-2: pantothenic acid-2
VIR. Right of rib-1 (<1 to 3%). Left of del (6%) and
trp-2 (11%) (140, 141,
143, 818, PB).
Unable to convert ketovaline to ketopantoic acid (138, 140, 141). Used in
major studies of intralocus recombination and complementation (140-143).
pan-2 ascospores remain white or pale if the crossing medium is not
supplemented, even when the
protoperithecial parent is pan-2+. Asci in which gene conversion has
occurred at pan-2 can thus be recognized and isolated (1072, 1073);
photographs (1072). For good recovery of pan-2 progeny, crossing media
should be supplemented with pantothenic acid (10 µg/ml) even when the
protoperithecial parent is pan+. Called group B.
pat: patch
IL. Linked to mt, probably to the right (1014).
Growth and conidiation occur in patches, in a cyclic pattern under certain
conditions (1014). Initially found in a pro-1 (21863) stock; pro-1 is
not
necessary for expression of patch. Original patch isolates were all sorbose
resistant (1014), but a sorbose-resistant derivative has been obtained that
apparently does not express the patch phenotype; this is called sor-4, q.v.
(816).
It is not clear whether pat and sor-4 are separate genes or whether
patch is not
scorable in the absence of modifiers present in the parent stock; see p. 267 of
reference 816. The original patch strain was used for the first demonstration
of a circadian rhythm in fungi (104, 829), but bd strains are now preferred.
pcon: phosphatase control
IIR. Right of the T(NM177) left breakpoint. Left of preg (1 to
2%) and pe
(4%). Probably allelic with nuc-2 (0/854) (593, 671).
Regulator gene of repressible alkaline phosphatase (671) and other steps in
phosphorus uptake and metabolism (665; R.L. Metzenberg, personal
communication). Constitutive allele pconc is dominant to, or codominant
with,
pcon+ (671). Scored on high-phosphate medium by staining reaction with
alpha-naphthyl phosphate plus diazo blue B (397, 1077). Used to study
phosphate transport (624). pconc allele c-6 called UW-6 in reference
593. See nuc-2. For regulation model, see references 665 and 670.
pdx-1: pyridoxine-1
IVR. Right of pyr-1 (<1 to 10%). Left of T(S1229) and
pt (2%) (40, 55, 692,
808). (482)
Uses pyridoxine, pyridoxal, or pyridoxamine (843, 845, 846). Shows intralocus
complementation (845, 846) and recombination (848), Provided the first proven
example of gene conversion (686). Scoring is sharpened by addition of 100 mg
of desoxypyridoxine per liter (845). Several alleles (called pdxp: e.g., 44602)
are pH sensitive and can grow without pyridoxine on
medium containing ammonium ions at a pH above 6 (1029). Conidia are
subject to death by unbaianced growth on minimal medium (1033). A yellow
pigment is excreted under certain conditions by the pdx-1;En(pdx) double
mutant; see En(pdx). Allele 44204 originally called pdx-2 (see
reference 848).
pdx-2
See pdx-1.
pe: peach
IIR. Between nuc-2 (4%) and arg-12 (1 to 5%) (593, 816).
(613)
Peach-colored conidia and short hyphae formed, more uniformly than by the
wild type, as a lawn close to surface of agar. Distinctive morphology (46, 613).
Added arginine increases macroconidiation and tends to obscure scoring of pe
at 25 C, but not at 39°C. pe single mutants produce both macro- and
microconidia. pe fl double mutants produce abundant grey microconidia and
no macroconidia (46, 700) (see fl). See col-1, col-4, and references
415 and
416 for interactions with other genes. Called m (microconidial) or
pem in
some contexts.
pen-1: perithecial neck-1
Unmapped.
Perithecia lack beaks (necks) when the pen-1 mutant is used as the female;
perithecia are normal when the pen-1 mutant is used as the male to fertilize
a pen+ strain (253).
per-1: perithecial-1
VR. Right of asp (26%) and at (8 to 14%). Left of ilv
(4%) (489, PB) and ts
(25%) (527).
Perithecial walls are devoid of black pigment when the female parent carries
per-1, regardless of genotype of the fertilizing parent (489, 490, 527). Alleles
are of two types (490). Type I produces young, completely white perithecia
that become pale yellowish after several days, and per-1 ascospores are white
(e.g., alleles PBJ1, ABT8, and AR174). Type II produces mature perithecia
that are somewhat darker orange with black pigment in the neck, and per-1
ascospores are normal black (e.g., alleles 29278, 29-281, and UG1837). Unlike
the perithecial wall trait, the ascospore trait shows no maternal effect. Black
pigment develops in a ring around the ostiole of type II perithecia, but is pale
or lacking in type I perithecia (490). Mosaic perithecia from heterokaryons
have been used for a clonal analysis of perithecial development (527, 528).
Expression is completely autonomous in ascospores (photographs in reference
529) and at least partially so in the perithecial walls (527-529). Used to test
for variegated-type position effect, with negative results (532). White per-1
ascospores (type I) germinate without heat shock and are usually killed by
hypochlorite or by the 30-min, 60°C treatment used to activate normal
ascospores (490, 527). Beaks of perithecia homozygous for allele PBJL (type
I) are abnormal, and ascospores are not shot properly (N.B. Raju, personal
communication). Type I alleles initially called sw: snow white (527).
Perithecial development mutants
See fs, ft, fmf, mb, mei, pen, and per.
Permease
See Transport.
Pf: Puff
IVR. Right of pyr-2 (2%). Linked to mat (3%) (812,
991).
Spreading colonial morphology (812).
pgov: phosphorus governance (provisional name)
IIIR. Linked to tyr-1 (1 to 4%), probably to the right (R.L. Metzenberg,
personal communication).
Regulatory gene for phosphorus uptake and metabolism. pgovc phenotype
similar to that of pregc (665; R.L. Metzenberg, personal communication).
Isolated in preg+/preg+ partial diploids. pgov allele c-5
is largely or
completely recessive in duplications from T(D305) (R.L. Metzenberg, personal
communication). Scored on high-phosphate medium by staining reaction with
alpha-naphthyl phosphate plus diazo blue B (397, 1077). For regulation
model, see references 665 and 670.
phe-1: phenylalanine-1
IL. Right of In(H4250) and suc (<1%). Left of ad-5
(816; H.B. Howe, Jr.,
personal communication). (48) [Duplications from In(H4250) x
phe-1 crosses
are phe-, unlike duplications from T(39311). The contrary
statement on p.
268 of reference 816 is a misprint.]
Originally reported to grow on phenylalanine, other aromatic amino acids,
leucine, or ethyl acetoacetate, with phenylalanine being most effective; several
other acids gave smaller responses (48). Utilization of phenylalanine and other
compounds varies for different isolates and on different carbon sources;
glycerol or ribose is preferable to sucrose (521-523, 753). Strains carrying
allele NM160 do not use phenylalanine but grow well on tyrosine or leucine
(816; A.G. De Busk, personal communication), at least with the strains and
carbon source used. The mutant phe-1 is inhibited by basic amino acids on
low-phenylalanine or leucine medium (48, 521). Growth on beta-labeled
leucine or beta-labeled phenylalanine showed that neither compound is
converted to the other (45). Called phen-1. Allele NM160 originally called
tyr(NM160) (316).
phe-2: phenylalanine-2
IIIR. Linked to vel (1%), between T(D305) and tyr-1
(2 to 4%) (40, 316,
809).
Lacks prephenic dehydratase (40, 316) (Fig. 11).
Requirement is very leaky.
Grows extensively and is treacherous to score by growth on minimal versus
supplemented medium, but can be scored reliably by blue fluorescence under
long-wave UV after growth on minimal medium without phenylalanine (816).
Appearance of phenylalanine in culture medium (118) is due to spontaneous
conversion of accumulated pretyrosine (519). Called phen-2. Allele Y16329
formerly called phen-3 (316).
phe-3:
See phe-2.
phen
Changed to phe.
pho-1: phosphatase-1
Possibly II. 20% wild-type recombinants with nuc-2. Independent of
nuc-1.
Low activity of the repressible alkaline phosphatase. Recessive. Complements
nuc-1 and nuc-2 in heterokaryons. Stains pale red on
low-phosphate medium
with alpha-naphthyl phosphate plus diazo blue B. (1077)
pho-2: phosphatase-2
VR. Between his-1 (3%) and inl (4%) (397; R.L.
Metzenberg, personal
communication).
Structural gene for repressible alkaline phosphatase (397, 594, 745). Scored
on low-phosphate medium by staining reaction with alpha-naphthyl phosphate
plus diazo blue B (397, 1077).
pho-3: phosphatase-3
IVR. Linked to pan-1 (<1%). Right of leu-2 (8%). Left of
mat (18%) and
the T(NM152) right breakpoint (745).
Structural gene for repressible acid phosphatase with phosphodiesterase activity
(745). Codominant in heterozygous pho-3/pho-3+ duplications.
Apparently
a member of both phosphorus and nitrogen regulatory circuits. Not under
control of the nit-2 locus (278b). Scored on low-phosphate medium by
staining
reaction with bis-nitrophenylphosphate (745).
pi: pile
IIL. Linked to ro-7 (0/75); left of cys-3 (4%)
(816).
Spreading growth (allele B101); no conidia. (816) Growth is better on minimal
medium than on complex complete medium. The mutant pi (B101) has not
been tested for 6-phosphogluconate dehydrogenase, which is deficient in
strain(s) carrying putative allele col-10 (R2438) (947). See col-10
regarding
allelism. As a marker, pi (B101) is preferable to col-10 (R2438)
because of
growth rate, stability, and ease of handling. ro-7, in the same region, is
preferable to both (PB).
pk: peak (synonym: bis, biscuit)
VR. Between met-3 (1%) and cot-2 (8%). Left of cl
(2%) (296, 818, 1036,
PB).
Initially colonial with flat surface growth; then sending up a mass of aerial
hyphae which conidiate profusely (789). Morphology somewhat similar to that
of sn, cum, sp, and cot-4 mutants at 25 C, but distinguishable.
Hyphae
branch dichotomously (713, 789). Asci are thin-walled, bulbous, nonlinear in
homozygous pk x pk crosses (714, 717, 792). Most alleles are
recessive for the
ascus effect, but some are dominant. Sorbose resistance mutations at various
loci act as dominance modifiers of the ascus effect of dominant alleles (898).
Increased activity of L-glutamine:D-fructose-6-phosphate amidotransferase was
observed in crude extracts of one pk strain but not in nine others; increased
activity for this enzyme was also found in cl (see below) and in four other
nonallelic morphological mutants (899). Hexoseaminoglycan consists of a
single component on medium without sorbose, in contrast to two components
for the wild type (1003). Antigenic surface mucopolyoside (281). Cell wall
analysis and photograph, allele B6 (278), allele C1810-1 (132). Cell wall
enzymes (334). Effect of carbon sources (280). One observation suggests
functional interaction (possible allelism) with cl (1007), but substantial
crossing-over frequencies and recovery of the pk-2 cl double mutant favor
separate loci (296). Several alleles have been previously called bis (for
nomenclature, see p. 270 of reference 816). Allele C-1610, originally called
pk-1, is inseparable from a reciprocal translocation, T(I;V)C-1610
pk (808).
Dominant allele 17-088 is associated with a V;VII translocation (A.M. Srb,
personal communication; PB).
pl: plug
VR. Linked to gran (0/75); between asn (1 to 9%) and
his-6 (16%) (812, 816,
1036).
Dense hyphae fill diameter of 10-mm tube (789). Morphologically distinct
from the mutant gran. Complex complete medium stimulates conidiation;
scoring of morphology is clearer on minimal medium.
pmb: permease basic amino acid
IVR. Right of uvs-2 (8%) (S. Ogilvie-Villa, cited in reference 248;
R. Sadler
and S. Ogilvie-Villa, personal communication).
Defective in basic L-amino acid transport (system III as defined in reference
778); reduced uptake of L-arginine, L-lysine, and L-histidine (778, 1152,
1076). Used extensively for transport studies; see reference 1150. Altered
surface glycoprotein (1038). pmb mutants selected as resistant to L-canavanine
(889, 1152). Allelic with bat (R. Sadler and S. Ogilvie-Villa, personal
communication), which was selected in arg-12s;pyr-3 (CPS- ACT+) by ability
to grow on minimal medium plus arginine, when the parental double mutant
was not able to grow because of arginine uptake and feedback onto the
arginine-specific carbamyl phosphate synthase (1074). Possibly allelic with
basa, which was selected by the inability of the mutant his-3 to grow
on
histidine plus methionine (628). Probably allelic with bm-1 (linked to
pyr-2,
24%), which was selected by canavanine resistance (913). Probably allelic with
argR (565, 566), q.v. Called Cr-10, Pm-B, pm b, UM-535, and
can-37. See
Transport.
pmg: permease general amino acid
Not mapped. Centromere linked. Not linked to mtr (pmn) or to
pmb in IVR
(248). (Previous report of mating type linkage [862] not confirmed).
Greatly reduced in general amino acid transport system II (as defined in
reference 777). Reduced uptake of arginine and phenylalanine. Selected by
resistance to p-fluorophenylalanine in a neutral (system I), basic (system
III) double mutant on medium lacking ammonium ions, where system II would
be derepressed in the wild type (248, 862, 863). A non-metabolizable substrate
specific for this transport system has been found (767). Called pm g. See
Transport.
pmn: permease neutral amino acid
Allelic with mtr (R. Sadler and S. Ogilvie-Villa, personal communication; A.G.
De Busk, personal communication), q. v. - Called Pm-N and pm n.
[pokyl: poky (synonym [mi-1])
Mitochondrial mutant with slow growth and deficient cyanide-sensitive
respiration (see reference 394). See su([mi-1]).
pp: protoperithecia
No data on linkage or alielism with already mapped loci that affect perithecial
formation or ascospore viability (e.g., gul-3, 4, -5; le-1, -2; ff,-fs)
Female sterile. Protoperithecia not formed. Ascospore lethal. Enhances
growth of the mutant gpi on glucose or sucrose. (711)
prd-1: period-1
III. Linked to acr-2 (5%), the centromere (0/35 asci), and pro-1
(20%) (327;
G.F. Gardner, personal communication).
Altered period of circadian conidiation rhythm. One allele is known, which
results in a 25.8-h period (at 25°C without csp) (327, 375). Recessive. Grows
at 60% of wild-type rate. See reference 327 for period lengths of double
mutants carrying frq-1, -2, and -3. Temperature compensation
described (377).
Name changed from frq-5 (375). For reviews of circadian mutants see
references 326 and 328.
prd-2: period-2
VR. (377)
Altered period of circadian rhythm (25.5 h at 25°C without csp for
allele
IV-2). Slower-than-normal growth. Recessive (329). Temperature
compensation described (377). Called IV-2.
prd-3: period-3
I. Near centromere (377).
Altered period of circadian rhythm (25.1 h at 25°C without csp for
allele IV-4)
(329). Slower-than-normal growth. Recessive. Temperature compensation
described (377). Called IV-4.
prd-4: period-4
Unmapped. Not allelic with frq or prd-1, -2, or -3 (329).
Altered period of circadian rhythm (18.0 h at 25°C without csp for allele
V-7)
(329). Normal growth rate. Dominant. Temperature compensation described
(377). Called V-7.
preg: phosphatase regulation
IIR. Right of nuc-2 (pcon) (1 to 2%), q.v. (671).
Regulator of repressible alkaline phosphatase and other steps in phosphorus
uptake and metabolism. Hypostatic to nuc-1, epistatic to nuc-2.
pregc
constitutive mutation is recessive to its wild-type allele. (665, 671; R.L.
Metzenberg, personal communication) Scored on high-phosphate medium by
staining reaction with (alpha-naphthyl phosphate plus diazo blue B (397, 1077).
Used to study phosphate transport (624). For regulation model, see
references 665 and 670.
pro-1: proline-1
IIIR. Between ser-1 (3/38 asci) (504) and ace-2 (1 to 9%) (578).
Right of sc
(7%) (814). (482)
Uses proline but not ornithine, citrulline, or arginine (1009). Structural gene
for pyrroline-5-carboxylate reductase (1177) (Fig. 10).
pro-3: proline-3 (synonym: arg-8)
VR. Linked to inl (0/74). Between his-1 (4%) and pk (2
to 6%). (812)
Uses proline, ornithine, citrulline, or arginine (1006). Blocked in the proline
pathway, in reduction of glutamic acid to glutamic-gamma-semialdehyde
(1105). Arginine and citrulline are used via the arginine catabolic pathway
(arginase and omithine transaminase), and ornithine is used via ornithine
transaminase (151, 234, 1104, 1105) (Fig. 10). Tends to
accumulate second
mutations, including arg-2 and his-1 (58, 994). Ability to grow on
arginine is
modified by ipa (994) and ota (234). Suppressed by
su(pro-3), which is allelic
with or closely linked to arg-6 (1129). Called arg-8.
pro-4: proline-4 (synonym: arg-9)
IIIR. Linked to thi-2 (0/78) and ota (4%) (818; D.J. West,
cited in
Neurospora Newsl. 16:19-22, 1970).
Uses proline, ornithine, citrulline, or arginine (1009). Proline pathway blocked
in reduction of glutamic acid to glutamic-gamma-semialdehyde (1105) (Fig. 10).
Citrulline and arginine are used via the arginine catabolic pathway (arginase
and ornithine transaminase), and ornithine is used via ornithine transaminase
(151, 1104, 1105). Leaky. Called arg-9.
prol
Changed to pro.
prt: protease
See pts.
psi-1: protein synthesis initiation-1
IVR. Right of the centromere (D.R. Stadler, A.M. Towe, and M. Loo, cited
in reference 619). Left of T(ALS159) and pyr-1 (4%) (808,
PB).
Conidial germination and hyphal growth inhibited at 35°C but normal (or
nearly so) at 20°C. Protein synthesis reduced after shift to restrictive
temperature. Recessive in heterokaryons. (619) Scored as an irreparable heat-
sensitive un mutant (see un).
pt: phenylalanine plus tyrosine
IVR. Right of T(S1229) and pdx-1 (2%). Left of col-4
(2%) (40, 55, 808).
(201) Original S4342 strain contained linked but separable insertional
translocation T(S4342) (808), the presence of which should not change
conclusions regarding gene order given in reference 40.
Requires phenylalanine plus tyrosine (201). Lacks chorismate mutase (40, 316)
(Fig. 11). Evidently the structural gene; strains carrying
allele NS1 have thermolabile chorismate mutase (D.E.A. Catcheside, personal
communication). NS1 strains are temperature sensitive, growing on minimal
medium at 25 C, where they are readily scorable by blue fluorescence under
long-wave UV and by browning of medium of aging cultures (1035). Inhibited
by complex complete medium.
pts-1: protease-1
Unmapped. Segregates as single gene not closely linked to alcoy markers (40
isolates) (441).
Structural gene for carbon-, nitrogen-, and sulfur-controlled extracellular
alkaline protease. Allele found in a single wild-collected strain, Groveland-1c
a, FGSC no. 1945. Synthesizes fast electrophoretic variant under conditions
of limiting carbon, nitrogen, and sulfur. Called prt (441). Regulation
reviewed
in references 642 and 665.
Purine
See ad and gua.
put-1: putrescine-1
Changed to spe-1, q.v.
pyr: pyrimidine
All pyrimidine auxotrophs of Neurospora are nonspecific, responding to any
pyrimidine nucleoside, nucleotide, or base. The symbol pyr is, therefore, used
for genes concerned with the biosynthetic pathway. Nucleosides or nucleotides
are more effective than corresponding bases as growth supplements for the
mutant pyr-1 (623) and apparently for other pyr mutants. However,
after a lag,
uracil is used nearly as effectively as uridine (683). No cytidine or
thymidine-specific requirement exists, because Neurospora lacks thymidine
kinase (421) and because any exogenous pyrimidine supplement is cycled back
through uridine monophosphate, which provides all the normal end products
of pyrimidine biosynthesis (1141). For this reason, DNA cannot be specifically
labeled by supplying [3H]thymidine, under normal circumstances. Mutations
have been obtained (uc-2, -3, 4, -5; ud-1) that block the pathway back through
uridine monophosphate and so prevent general labeling from a single precursor
(1141). Cytosine in DNA can be labeled specifically by the method of Worthy
and Epler (1163). For a general review of pyrimidine metabolism, see
reference 766. For systematic gene-enzyme work, see references 133 and 134.
For pyrimidine biosynthetic pathway, see Fig. 20. For loci
concerned with
pyrimidine salvage or pyrimidine uptake, see uc, ud, udk, and Fig. 23. Complex interactions between lys and
pyr mutations have been
described
(485).
Pyrimidine biosynthetic enzymes differ in their modes of regulation. The
pyrimidine-specific carbamyl phosphate synthase-aspartate carbamyl transferase
complex is derepressed by end product depletion, but is insensitive to
repression in the fluoropyrimidine-resistant mutant fdu-2 (127, 135);
dihydroorotase is relatively unresponsive to end-product limitation;
and dihydroorotate dehydrogenase is induced by a precursor which is probably,
by analogy with Saccharomyces, dihydroorotate, the substrate of the enzyme.
Regulation of the last two enzymes has not been studied systematically.
Pyrimidine regulation of the uptake and salvage pathways of pyrimidine is
discussed under individual loci; see uc-5 and ud-1. Many aspects of
pyrimidine
metabolism are under the control of general nitrogen metabolite regulation
(128).
pyr-1: pyrimidine-1
IVR. Right of psi (4%) and T(ALS159) (808, PB). Left of
pdx-1 (<1 to 10%)
(692). (482)
Requires uracil or other pyrimidine. Lacks dihydroorotate dehydrogenase
activity (133, 134) (Fig. 20). All ascospores from
pyr-1 x pyr-1
crosses are
white if cross is on medium containing 0.1 mg of uracil per ml; they are
black if the cross is on medium containing 1.0 mg/ml (632).
pyr-2: pyrimidine-2
IVR. Right of nit-3 (2 to 9%). Left of mat (3%) and the
T(NM152) right
breakpoint (633, 812, 1000, PB). (482)
Requires uracil or other pyrimidine. Lacks orotidine
5'-monophosphate pyrophosphorylase activity (133, 134) (Fig.
20). Needs
medium containing >0.5 mg of uracil per ml for optimal growth. Allele 38502
is leaky.
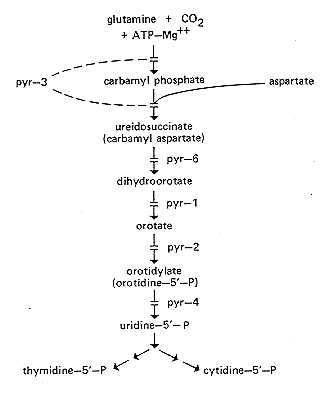
FIG. 20. Pyrimidine biosynthetic pathway, showing sites of gene action (134,
456, 841, 1140). Carbamyl phosphate for arginine synthesis is made as a
separate pool by a different enzyme system (see arg-2, arg-3, Fig. 10).
Interchange between the two pools occurs only in certain mutant combinations.
ATP, Adenosine 5'-monophosphate.
pyr-3: pyrimidine-3
IVR. Right of the T(NM152) left breakpoint and of T(S1229);
hence, right of
arg-14. Left of his-5 (1%) (238, 808). (482)
Requires uracil or other pyrimidine (683). Growth inhibited by purine
nucleosides and nucleotides (825). Structural gene for pyrimidine-specific
carbamyl phosphate synthase (CPS) and aspartate carbamyl transferase
(ACT; also abbreviated ATC) (456, 850) (Fig. 20). Mutants
may lack either
or both activities, e.g., those carrying alleles KS43 (CPS+ ACT-), KS20 (CPS-
ACT+), and KS11 (CPS- ACT-) (1140). Unlike Saccharomyces, no
feedback-insensitive CPS+ ACT+ mutants of Neurospora have been
discovered (A. Radford, unpublished data). Some mutants have kinetically
altered aspartate carbamyl transferase (456, 880). Used extensively for studies
of channeling and relation of gene structure to the two enzyme activities (236).
Normally, carbamyl phosphate produced by pyr-3+ is used solely for
pyrimidine synthesis, and carbamyl phosphate produced by arg-2+ and
arg-3+
is used for arginine synthesis, the enzymes being in different organelles;
however, a deficiency of the next enzyme in either pathway permits overflow
of carbamyl phosphate into the other pathway (reviewed in reference 236).
Hence, CPS- ACT+ alleles are suppressed by arg-12s (246), and CPS+' ACT-
alleles can be selected as suppressors of arg-2 and arg-3 (658, 887,
and
references therein). Some of the CPS+ ACT- mutations, called pyr-su-arg,
suppress the arginine requirement but retain enough aspartate carbamyl
transferase activity that they have no detectable pyrimidine requirement (877,
881). arg-13, arg-4, arg-5, arg-6, and am partly suppress CPS- ATC+
alleles
(see reference 660). Fine-structure map (851, 1050). Fertility of interallelic
crosses is variable and often very poor (658). Complementation between CPS-
ACT+ and CPS+ ACT- mutants (246) and between some pairs of CPS+
ACT- mutants is good; otherwise, complementation is poor (849, 1159).
Complementation maps (658, 849, 877, 1159). Mutational analysis (852).
Direction of translation, based on enzyme types of polar mutants, is from CPS
to ACT (850). Allele 37815(t) is heat sensitive (34°C versus 25 C) (68). Allele
1298 is C02 remediable (191, 192). Strain KS12, a pyr-1 pyr-3 double
mutant, was originally called pyr-5 (see reference 346). The different classes
of pyr-3 alleles have been called M (CPS-P-less), N (ACT-less), and MN
(lacks
both activities).
pyr-4: pyrimidine-4
IIL. Right of het-c (1%), T(P2869), and cys-3 (18 to
21%). Left of ro-3 (1 to
2%) (721, 816, PB). (812).
Requires uracil or other pyrimidine. Lacks orotidine 5'-monophosphate
decarboxylase (133, 134, 841) (Fig. 20). Fertile crosses
homozygous for pyr-4
can be made by using very high levels of uridine (15 to 20 mg/ml) (O.M.
Mylyk, personal communication).
pyr-5: pyrimidine-5
A pyr-1 pyr-3 double-mutant strain was originally called pyr-5 (346).
pyr-6: pyrimidine-6
VR. Between asn (6%) and un-9 (2%) (156, 818). (133,
134)
Requires uracil or other pyrimidine. Lacks dihydroorotase activity (133, 134)
(Fig. 20). On a small amount of uridine, a strain(s) carrying the
only allele
(DFC37) grows, after a pause, well beyond the level normally supported by the
supplement; at no time is dihydroorotase detectable (134).
q: quinolinic acid
See nic-1.
qa: quinate utilization
Gene cluster located in linkage group VII (177, 884), in the order: qa-1
(0.02%) qa-3 qa-4 qa-2 (0.02%) met-7 (146). No recombination
with the
centromere has been observed in several hundred asci; qa has been tentatively
shown right of the centromere, on the basis of close linkage to met-7 (146;
M.E. Case, personal communication). The gene cluster functions in the quinate
catabolic pathway. Mutants selected by inability to use quinate as their
sole source of carbon (178, 179, 885). The enzymes of the quinate pathway are
induced when quinate is present in the medium; but see also reference 423 and
references therein. Scored by the ability to use quinic acid (0.3%)
as the sole carbon source (146). qa-1 encodes a regulatory protein; the other
qa genes encode inducible enzymes (179). The first and second steps of the
quinate pathway are paralleled by comparable reactions in the aromatic
biosynthetic pathway (Fig. 11). Thus, the enzymes encoded
by aro-9 and
qa-2
can substitute for each other, and under appropriate conditions the qa-3
enzyme can substitute for the aro-1 enzyme (147). Separate transcripts are
made by components of the qa cluster, rather than a single polycistronic
messenger RNA (387, 781). For a diagram of the catabolic pathway and its
relation to the biosynthetic pathway, see references 146, 387, and 423 and
Fig. 21. The entire cluster has been cloned in E. coli
and returned to
Neurospora by transformation (941). The cluster consists of adjacent coding
sequences totaling about 18 kilobases. A transcriptional map has been
constructed, and two new genes of unknown function, qa-x and qa-y,
have been
identified from messenger RNAs (781). For regulation, see references 179,
423, 781, and 873.
qa-1: quinate-1
VII. Left of met-7 (0.2%). Leftmost gene in the qa cluster (146).
(884)
Unable to use quinate or shikimate as the sole carbon source. Quinate
catabolic pathway regulatory gene (179). qa-1 mutants (recessive) are
deficient in all three enzymes of the pathway: quinate dehydrogenase
(shikimate dehydrogenase), catabolic dehydroquinase, and dehydroshikimate dehydrase
(Fig. 21) (885). qa-1F and qa-1s are fast-
and
slow-complementing alleles which appear to define two
nonoverlapping segments of the gene. Strains carrying qa-1c alleles are
constitutive producers of the three enzymes; these are readily
found among revertants of qa-1 (145a, 178, 387). Expression of
qa-1 appears
constitutive but also autoregulated (781).
qa-2: quinate-2
VII. In the qa cluster between qa-4 and met-7 (0.02%)
(146). (884)
Lacks catabolic dehydroquinase (148, 517, 884). Unable to use quinate
or shikimate as the sole carbon source in the presence of aro-9,
which results in the absence of biosynthetic dehydroquinase (885) (Fig. 21).
aro-9;qa-2+ strains grow on minimal medium without supplement.
qa-2 is
conveniently scored as an aro auxotroph when aro-9 is present.
qa-2+ cloned
in pBR322 (pVK57) is expressed constitutively from its own promoter in E.
coli
(23, 388, 451); has been returned to Neurospora chromosomal sites by
transformation (150). Allele M246 is stable (150).
qa-3: quinate-3
VII. In the qa cluster between qa-1 and qa-4 (146).
(177)
Unable to use quinate or shikimate as the sole carbon source (178). Structural
gene for quinate (shikimate) dehydrogenase (178) (Fig. 21).
Revertants have
altered enzymes. Transcribed in the direction qa-4 to qa-1 (149).
qa-4: quinate-4
VII. In the qa cluster between qa-3 and qa-2 (146).
(177)
Unable to use quinate or shikimate as the sole carbon source (178). Lacks
dehydroshikimate dehydrase (178) (Fig. 21).
qa-x, qa-y
See qa.
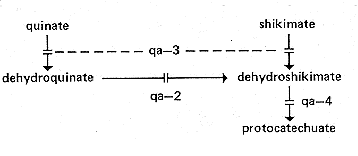
FIG. 21. Quinate (aromatic amino acid) catabolic pathway, showing sites of
gene action (146, 148, 387, 423, 517, and references therein). qa-1 is a
regulatory gene affecting all three structural genes. qa-2+ activity (catabolic
dehydroquinase) can be replaced by the product of aro-9+, the equivalent
gene
in the aromatic biosynthetic pathway. See reference 423 for the catabolic steps
subsequent to protocatechuic acid.
R: Round spore
IR. Right of T(NM169d) and aro-8 (4%). Left of un-18
(11%) (1093). (H.R.
Cameron, personal communication)
All eight ascospores of heterozygous R/+ asci are round rather than ellipsoid.
R is thus nonautonomous in ascospores and dominant in the ascus (690).
Ascospores are round even in nonlinear asci (1008; D.D. Perkins, unpublished
data). Usually two germination pores are formed, but sometimes one (1008).
Photograph (1008). Vegetative morphology abnormal, somewhat resembling
that of pe mutants. Initial growth on slants is concentrated around the
inoculation point. The vegetative morphology is recessive in heterozygous
duplications, as from T(NM103) (1091). Female sterile, with no perithecia,
but
R x R crosses can be accomplished if R is heterokaryotic
in the female parent
(A.M. Srb, personal communication). Used in studies of duplication instability
(1091) and autonomy of ascospore development (529). Allelic
recurrences obtained (1008; A.M. Srb, personal communication). For other
genetically determined round ascospores, see reference 59. Called Rsp (1008),
but the original symbol R has priority. (rsp has been used for
cytoplasmically
determined respiration-deficient mutants [890].)
r(Sk-2)-1: first locus resistant to Spore killer-2
IIIL. Left of acr-7 (7%) and sc (17%) (1092; B.C.
Turner, personal
communication). Allelism of r(Sk-2)-1 with Sk-2 is not excluded
because
Sk-2K blocks recombination in this region. r(Sk-2)-1 itself does not
block
recombination.
Prevents killing of ascospores by Sk-2K - Not resistant to killing by
Sk-3K.
Allele P527 found in N. crassa from Louisiana. Only one other known N.
crassa wild type is resistant. (B.C. Turner, personal communication)
Radiation sensitive
See mus, uvs. See also gs, Mei-2, mei-3, nuh-4, and
upr-1. The symbol rad has
not been used in Neurospora.
rDNA: ribosomal DNA
Used to designate genes specifying 5.8S 17S, and 26S rRNA, which are located
in tandemly repeated units in the nucleolus organizer region. See NO.
rec: recombination
A class of genes affecting meiotic recombination in specific loci or regions
(175, 520). Initially detected by changed intralocus recombination (up to 25x),
but interlocus crossing over may also be affected (up to 40X). Polarity of
intralocus recombination may be changed or reduced by the presence of rec+.
Any given target locus or region appears to be affected by alleles at only one
specific rec locus (Fig. 22). High recombination is
recessive. Products of
dominant alleles (called rec+) are thought to repress initiation of
recombination at specific recognition (cog) sites by binding to an adjacent
con
(control) site. Control of recombination is independent from control of gene
expression (153). Allelic differences are present in commonly used laboratory
wild types (174). For reviews, see references 167, 169, 170, and 343.
rec-1: recombination-1
VR. Between ro-4 (7%) and asn (5%) (159). (165)
Presence of allele rec-1+ reduces recombination within the loci
his-1 (VR)
(520, 1070) and nit-2 (IL) (155, 157) (Fig. 22). A
recessive allele
from another lineage was called rec-z until probable identity with
rec-1 was
established (157). rec-1+ does not affect recombination within any other
his
locus tested (172).
rec-2: recombination-2
VR. Between sp and am (174). (993)
Presence of dominant allele rec-2+ reduces recombination within the
his-3
locus (IR); also reduces crossing over in the intervals pyr-3-his-5
(IVR), his-3-ad-3 (IR), and arg-3-sn (IL) (171, 74, 992) (Fig. 22).
Interacts
with cog in affecting recombination in his-3 and crossing over
between his-3
and ad-3 (27, 171). Used in conjunction with translocation TM429
to
demonstrate the cis action of cog+ on recombination between sites
in his-3
(171). (See cog.) Recessive rec-3 alleles from other lineages were
called rec-4,
rec-5, or rec-w until identity was demonstrated (see reference 167).
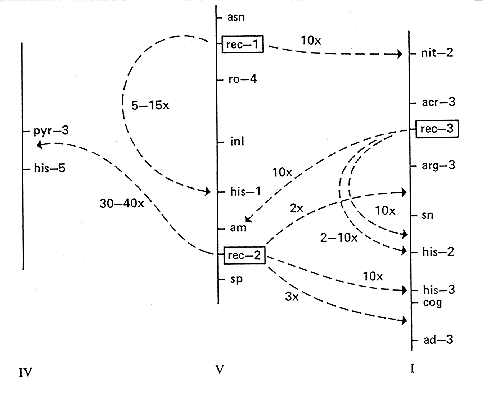
FIG. 22. Map locations of the rec genes. Arrows show the sites
where they
are known to affect meiotic intra- or interlocus recombination frequencies, and
the magnitude of the effect (170 and references therein; D.E.A. Catcheside,
personal communication).
rec-3: recombination-3
IL. Between acr-3 (1 to 2%) and arg-3 (2 to 6%) (168, 173).
(166)
Presence of allele rec-3+ reduces recombination within the loci
his-2 (IR) and
am (VR) but not in adjoining regions or within gul-1, which is less
than 0.3%
from am (173, 997, 998). Crossing over is also reduced in the interval
between
sn and his-2 (174) (Fig. 22). Three alleles
known: rec-3,
rec-3L, and rec-3+
(168). A recessive allele from another lineage was earlier called rec-x (see
reference 167).
rec-4, rec-5, rec-w
See rec-2.
rec-x
See rec-3.
rec-z
See rec-1.
rg-1: ragged
IR. Right of T(AR173); hence, of his-2. Left of lys-4 (1
to 7%) (271, 789, 810).
Spreading dense colonial growth with poor conidiation (789). Increased hyphal
branching; bumpy mycelial surface. Altered phosphoglucomutase (isozyme I).
Accumulates glucose-1-phosphate (117). Cell wall composition (132). Normal
levels of NADPH (110) and linolenic acid (115). Photograph (112). The
double mutant rg cr grows as small discrete conidiating colonies suitable for
velvet replication (634). For examples of applications, see references 932 and
1020. Unlike sn cr, which it resembles phenotypically, the double mutant
rg
cr is not homozygous fertile. Allele R2357 was formerly called er: erupt
(see
reference 382). Allele S4357 was formerly called col-7 (see reference 675).
rg-2: ragged-2
I. Linked to mt (15%) after introgression into N. crassa (D.D.
Perkins,
unpublished data). Interpreted as unlinked to rg-1 and su(rg-2) in
N. sitophila
(680).
Found in N. sitophila crosses involving an introgressed rg-1.
Morphology
similar to that of rg-1 mutants (680). Altered phosphoglucomutase (isozyme
II) (678).
rib: riboflavin
Only two rib loci are known in Neurospora, compared to six which
have been
assigned to biosynthetic steps in Saccharomyces (see reference 256).
Supplemented medium should be shielded from light to avoid destruction of
riboflavin.
rib-1: riboflavin-1
VIR. Between T(AR209) and pan-2 (3%). Right of
ad-1 (3 to 6%), the
centromere (1%), and glp-4 (4%) (486, 1012, 1102). (482)
Requires riboflavin (681). Used to demonstrate role of flavin as a
photoreceptor for carotenogenesis and for phase shifting and suppression
of circadian conidiation (775). Allele 51602 is heat sensitive (34°C versus 25
C); allele C106 is not (380).
rib-2: riboflavin-2
IVR. Right of T(4342). Left of chol-2. Probably left of
pyr-3 (1/24 asci) (381,
808).
Requires riboflavin (381). Used to demonstrate the role of flavin as a
photoreceptor for phase shifting of circadian conidiation and carotenogenesis
(775).
ribosomal RNA
Genes specifying 5.8S, 17S, and 26S rRNA (but not 5S) are located in the
nucleolus organizer region. See NO.
rip-1: ribosome production-1
IIR. Linked to un-15 (1%); right of fl and trp-3 (6 to
9%). (PB).
Conditional defect in production of 60S ribosomal subunits (622). At the
restrictive temperature (37 C), RNA synthesis is affected first and then protein
synthesis; 60S cytosolic ribosomal subunits are underaccumulated, and
relatively little 25S rRNA is produced (618, 620, 622; P.J. Russell, personal
communication). Defective ribosome biosynthesis at high temperatures is
attributed to a defect in rRNA processing (622). Attains 2.4% of the wild-type
growth rate at 35°C and 80% at 25°C (P.J. Russell, personal
communication).
Scored as an irreparable heat-sensitive un mutant (see un). Good
fertility,
growth, and viability make rip-1 preferable to un-15 as a marker for
the right
end of II. The onginal strain carrying both rip-1 and inl (89601)
(382, PB).
was called 4M(t). The rip-1 allele in this strain was originally called 1(t).
ro-1: ropy-1
IVR. Linked to pan-1 (0/394) (633).
Cable-like aggregates of hyphae grow up the tube from agar slants. Conidia
form in dense clumps at the top (789). Hyphae are curled microscopically
(382). Cell wall analysis; photograph (278). Reduced amount of cell wall
peptides (1165). Growth limited on glycerol medium (212). Fertile as the
male; perithecia rare or absent when used as the female.
ro-2: ropy-2
IIIR. Right of trp-1 (2 to 14%). Left of T(D305) and
phe-2 (5%) (11, 812,
814; D.D. Perkins, unpublished data).
Resembles ro-1 (789).
ro-3: ropy-3
IIL. Right of pyr-4 (1 to 2%). Left of T(NM149) and
thr-2 (6 to 25%) (808,
812).
Resembles ro-1 (812). Growth limited on glycerol medium (212). Called
cfl
on map in reference 812.
ro-4: ropy-4
VR. Linked to pab-2 (0/407). Between ad-7 (4%) and
inv (5%). (156, 382,
816, PB)
Resembles ro-1. Growth limited on glycerol medium (212). Mutations R2428
and R2520, called ro-5 and ro-8 in references 382 and 698, are
allelic with ro-4
allele B38 (816). Reduced amount of cell wall peptides (1165).
ro-5
Allelic with ro-4, q.v. (see reference 816).
ro-6: ropy-6
IR. Between the T(4540) breakpoints; hence, between nic-2 (0/95)
and thi-1
(382, PB).
Resembles ro-1 (382)
ro-7: ropy-7
IIL. Linked to pi (0/75). Left of cys-3 (11%) (382,
PB)
Resembles ro-1 (382). Growth limited on glycerol medium (212). Female
sterile, contrary to misprint in reference 811.
ro-8
Allelic with ro-4, q.v. (see reference 816).
ro-9: ropy-9
II. Right of T(NM149). Probably left of arg-5 (8%). Linked to
thr-3 (0/63)
(382, PB).
Growth limited on glycerol medium (212). Misnamed. Hyphae not curled,
unlike those of strains carrying other ro genes (D.D. Perkins, unpublished
data). Makes barren perithecia in the cross of allele R2526 x tng, which is
possibly allelic, but R2526 strains are poor female parents. Conidia of R2526
strains are normal sized, unlike those of the mutant tng.
ro-10: ropy-10
IL. Left of fr (18%) (PB).
Resembles ro-1 (PB)
rol-1: ropy-like-1
IV. Linked to pdx-1 (0/88) (382).
Resembles ropy strains in growth habit on slants, but hyphae are not curled
microscopically (382).
rol-2: ropy-like-2
VII. Linked to met-7 (0/298) (PB). (382)
Resembles rol-1 (382).
rol-3: ropy-like-3
VR. Between ilv-1 (2%) and cot-4 (5%) (698).
Resembles rol-1. Photograph: Fig. 16 in reference 382.
ros: rosy
Allelic with al-3, q.v. (PB).
Strains carrying al-3 allele Y234M70, called ros, produce pale pink
carotenoids more abundantly than strains carrying al-3 allele RP100 or
P7775 (PB). Original from T. Ishikawa. Studied briefly by A. M. Kapular.
Rsp: Round spore
See R.
Return to the FGSC Home page

Contact the FGSC
Last modified 4/24/96 KMC



