Neurospora mutants beginning with U through Z
You may jump directly to the following sections:
Unknown temperature sensitive (un)
Urease (ure)
UV sensitive (uvs)
White collar (wc)
Yellow (ylo)
uc: uracil salvage or uracil uptake
Symbol used in references 808b and 810 for mutations affecting the thymidine salvage
pathway (uc-1, -2, -3, -4) or transport of pyrimidine
bases (uc-5) (Fig. 23). Not used for biosynthetic
pathway mutants (see pyr for uracil biosynthesis).
uc-1: uracil salvage-1
II. Linked to pyr-4 (31%) (1139, 1141).
Altered thymidine salvage pathway (Fig. 23). Able to use
thymidine, thymine,
5-hydroxy-methyluracil, or 5-formyluracil as the sole pyrimidine source in
germinating conidia of pyr-4 mutants, which are blocked in pyrimidine
biosynthesis. (In uc-1+ background, pyr-4 can use these compounds
only
if a primer of uridine or cytidine is supplied [1141]). Causes elevated activities
of enzymes which oxidatively demethylate thymine (420).
uc-2: uracil salvage-2
I. Linked to mt (0/12 asci) (1139, 1141).
Thymidine salvage pathway defect (Fig. 23). Unable to use
thymidine or
deoxyuridine, but can use thymine, 5-hydroxymethyluracil, 5-formyluracil,
uracil, or uridine as the sole pyrimidine source for pyr-4 when uc-1
is also
present (1141). Reduced activity of the 2'-hydroxylase
reactions thymidine -> thymine ribonucleoside and deoxyuridine -> uridine
(977).
uc-3: uracil salvage-3
Not mapped.
Thymidine salvage pathway defect (Fig. 23). Unable to use
thymidine,
thymine, or hydroxy-methylcytosine, but can use deoxyuridine, 5-formyluracil,
uracil, or uridine to support growth of pyr-4 when uc-1 is also
present. Excretes
thymine into the medium when grown in the presence of thymidine (1141).
Lacks thymine 7-hydroxylase, which catalyzes the reactions
thymine -> 5-hydroxymethyluracil -> 5-formyluracil -> uracil-5-carboxylic acid;
apparently, there is an alternate enzyme for 5-formyluracil ->
uracil-5-carboxylic acid, which explains the growth on 5-formyluracil (616, 977).
uc-4: uracil salvage-4
VR. Between inl (12%) and his-6 (11%) (126). Not in I as
reported in
references 1139 and 1141 on the basis of one ascus in eight recombinant with
mt.
Thymidine salvage pathway defect (Fig. 23). Unable to use
thymidine,
thymine, 5-hydroxymethyluracil, 5-formyluracil, or uracil to support
pyr-1, even in the presence of a uridine primer (1141). Deficient in
phosphoribosyl transferase (127). The uc-4 single mutant converts 50%
of the supplied uridine to uracil and excretes it into the medium (1141).
Resistant to 5-fluorouracil (126).
uc-5: uracil uptake-5
IV. Right of cot-1 (32%) (126). (1139, 1141).
Apparently defective in transport of pyrimidine bases (Fig. 23).
Unable to use
any free pyrimidine base to support the growth of the pynmidine auxotroph
pyr-1, although it can use both ribose- and deoxyribose-nucleosides (1141).
Resistant to 5-fluorouracil (126). The pyr-1 uc-5 double mutant has been used
to study uptake inhibition of structural analogs (225). Selected by inability to
use uracil to supplement pyr auxotrophs, or as 5-fluorouracil resistant on
ammonia-free minimal medium (126). Uracil uptake is decreased by NH3
(629) or by other good nitrogen sources, and by a nit-2 mutation (128). Called
tru.
ud-1: uridine uptake-1
VIIR. Between met-7 (27%) and arg-10 (10%) (126).
(463).
Unable to use pyrimidine nucleosides to support the growth of a pyrimidine
auxotroph (pyr-1) although any free pyrimidine base can be used. Probably
defective in pyrimidine nucleoside transport (1141) (Fig. 23).
Apparently also
defective in purine nucleoside transport (224). Resistant to
5-fluorodeoxyuridine and 5-fluorouridine (126). Resistance is recessive in
heterokaryons. Shows interallelic complementation (127). Scored by spotting
conidial suspension on medium containing 4 x 10(-5)M 5-fluorodeoxyuridine,
filter sterilized (463). Mutant gene CIF-dUrd 7, selected by resistance and
called fdu-1 (463), is allelic (126). Uridine uptake is decreased by good
nitrogen sources and by nit-2 (128).
udk: uridine kinase
VR. Left of uc-4 (29%) (126).
Deficient in uridine kinase (127) (Fig. 23). The udk
uc-4 double mutant is
resistant to 5-fluoro-deoxyuridine and 5-fluorouridine (126), but the udk single
mutant is not resistant to any analog.
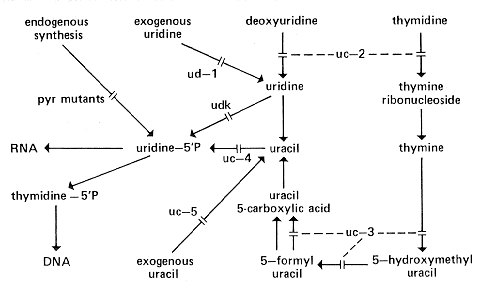
Fig. 23. Uracil salvage pathway, by which many exogenously supplied
pyrimidines are
cycled via uracil to uridine 5'-phosphate and, thus, into the latter part of the pyrimidine
biosynthetic pathway. The figure shows the sites of action of the salvage pathway
uc genes, the ud and uc-5 genes, which control uptake of
exogenous uridine and uracil, and udk, which controls uridine kinase (126, 420,
616, 977, 1141). For endogenous pyrimidine synthesis, see Fig. 20.
ufa-1: unsaturated fatty acids-1
IV or V. Linked to inl in a cross with alcoy (944). Data do not
distinguish
linkage group because T(IVR;VR)R2355 was present.
Requirement satisfied
by C16 or C18 fatty acids having a double bond in cis at either the Delta9 or
Delta11 position, by C16 fatty acids with double bond in trans at Delta9, or by
fatty acids with multiple cis double bonds interrupted by methylene bridges
(944, 945). Reverts readily, and revertants overgrow culture if grown on
suboptimal supplement (S. Brody, personal communication). Tween 80 (0.1%)
is satisfactory for maintenance. Stock viability is better on Tween 80 than on
fatty acids suspended in detergent Tergitol Nonidet P-40 (944). For
biosynthetic pathway see reference 944.
ufa-2: unsaturated fatty acids-2
IV or V. Linked to inl in a cross with alcoy (944). Data do not distinguish
linkage group because T(IVR;VR)R2355 was present.
Requirement similar to that of ufa-1 mutants. Linkage similar to that of
ufa-1.
No intercross data. Designation as second ufa locus is based solely on
complementation with ufa-1 (944). Highly revertible, and stocks are readily
lost on suboptimal medium (S. Brody, personal communication).
un: unknown
Unknown function. Temperature-sensitive (heat-sensitive) conditional mutants,
irreparable by supplementation at the restrictive temperature (usually 34 to 37
C). Originally referred to as "unknown requirement" on the initial hypothesis
that such mutants would prove to be auxotrophs. Several heat-
sensitive genes have been sufficiently characterized to be assigned more
specific names than un (e.g., ndc, rip, psi, eth-1, and fs-2).
At least some un
mutants are deficient in amino acid transport (543, 1075). For most un loci,
however, little is known of the molecular or cellular basis. Because of their
map locations, several are useful as genetic markers. Scoring may require
growth comparisons at two temperatures, preferably with small conidial
inocula. Some heat-sensitive mutants with altered morphology at restrictive
temperature are called cot or scot.
Temperature-sensitive auxotrophs with certain requirements have sometimes
been classed initially as un because complex complete medium is either
inadequate or inhibitory. Thus, complex-medium-irreparable
temperature-sensitive mutants no. 3, 13, and 14 of reference 507 proved to be
temperature-sensitive thr auxotrophs; no. 6, 20, and 30 were his; and
no. 19,
24, and 35 were asn (T. Ishikawa, personal communication). Many
un mutants
do not achieve normal growth rates even at the permissive temperature
(usually 25 C). Genes un-1 to un-8 were assigned locus numbers in
reference
813. "T" in allele numbers of several un mutants designates Tokyo, not
translocation.
un-1: unknown-1
IR. Between nic-2 and the T(4540) right breakpoint. Linked to
cys-9 (0/72),
cr-1 (5 to 9%), and bs (9%) (816, 818, PB). (482).
Unknown function. Heat sensitive. Growth at 25°C but not at 34°C (484).
Formerly called un(44409).
un-2: unknown-2
IR. Between the T(AR173) left breakpoint and T(AR190); hence,
right of the
centromere and arg-3 (1 to 2%) and left of his-2 (1%). Included in
duplications from T(AR173) but not from T(AR190) (808).
(482).
Unknown function. Heat sensitive (482). Growth at 25°C but not at 39°C.
Scorable but leaky at 34°C. Formerly called un(46006).
un-3: unknown-3
IL. Right of In(NM176); hence, right of ser-3 (1%) (1093). Left of
mt (0.04
to 0.1%) (488, 758). Closest bracketing marker left of mt (482).
Unknown function. Heat sensitive (484). Growth drops off sharply between
28.5°C and 30°C (R.L. Metzenberg, personal communication). Multiply
transport deficient at permissive temperatures with increased fragility of
protoplasts (543); reduced rate of uptake of citrulline (1075) and aspartate
(543, 1149). Resistant to ethionine and to p-fluorophenylalanine at 25°C (542,
543). Used to tag mating type (487) and as a flanker in an attempt to resolve
the mating type region by recombination (758). Strains with probable un-3
alleles are selected as citrulline-resistant mutants of pyr-3 arg-12s; most
mutants selected in this way show complementation between alleles (1075).
A possible functional relation to mating type is discussed in reference 543,
Growth at 25°C aided by 0.3 mg of sodium acetate per ml. May be scored by
slow growth at 25°C if acetate is not added to minimal medium (487).
Formerly called un(55701).
un-4: unknown-4
VIL. Right of lys-5 (2%). Left of T(T39M777) and cys-2
(4%) (808, 1012).
(482).
Unknown function. Heat sensitive, 34°C versus 25°C (481). Formerly
called
un(66204).
un-5: unknown-5
IL. Right of fr (6%). Left of T(39311) and nit-2 (2%)
(816, 798). (574).
Unknown function. Heat sensitive, 34°C versus 23°C. Inhibited by
histidine
and tryptophan: osmophilic (26 to 28 C) (574, 575). Formerly called un(b39).
un-6: unknown-6
IIIR. Right of sc (21%) and acr-2 (6 to 20%). (816, PB).
Unknown function. Heat sensitive, 34°C versus 25°C (484). Formerly
called
un(83106).
un-7: unknown-7
IR. Left of al-1 (3%) (818, 813). Between T(STL76) and
T(4637); hence, right
of cyh-1 (808).
Unknown function. Heat sensitive, 34°C versus 25°C (506). Dies slowly at
restrictive temperature (508). Allele T35M50 is called 31 or TS31 in
references 507 and 508.
un-8: unknown-8
IVR. Right of T(ALS159); hence, right of psi-1. Left of
col-4 (5%) and
T(S1229). Linked to pyr-1 (0/47). (813, PB).
Unknown function. Heat sensitive (507). No growth at 34°C. Morphology
abnormal at 25 C, unlike that of strains carrying closely linked psi. Allele
T27M9 is called 1 in reference 507.
un-9: unknown-9
VR. Between pyr-6 (3%) and his-6 (5 to 9%) (818).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T54M96 is
called 42 in reference 507.
un-10: unknown-10
VII. Right of wc-1 (7%). Left of frq (9%) and for
(12%) (818; J.F. Feldman,
personal communication).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T42M45 is
called 11 in reference 507.
un-11: unknown-11
VR. Linked to al-3 (0/48) (818).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T42M30 is
called 10 in reference 507.
un-12: unknown-12
IVR. Linked to col-4 (0/73) and pdx-1 (5%), left of
T(S1229) (818, PB).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T51M118 is
called 17 in reference 507.
un-13: unknown-13
Linked to ylo-1 (2%), un-4 (3%), and lys-5 (4%)
(818).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T42M24 is
called 9 in reference 507.
un-14: unknown-14
IIIR. Right of acr-2 and thi-4 (8 to 20%). Left of leu-1
(5%) (818, PB).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T54M55 is
called 36 in reference 507.
un-15: unknown-15
IIR. Linked to rip-1 (1%). Right of trp-3 (10%). (813,
PB).
Unknown function. Heat sensitive, 34°C versus 25°C (T. Ishikawa, personal
communication). Grows poorly at the permissive temperature; rip-1 is,
therefore, preferred to un-15 as a marker for the right end of II (811). One
allele, T54M50.
un-16: unknown-16
IL. Right of mt (<1%). Left of ta (1%) and acr-3
(<3%). Closest flanking
marker right of the mating type locus (818, PB).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T42M69 is
called 16 in reference 507.
un-17: unknown-17
IIIR. Between tyr-1 (4%) and dow (23 to 28%) (818).
Heat sensitive, 34°C versus 25°C (507). Ascospores containing
un-17 are white
or slow to mature (D.D. Perkins, unpublished data). Shows rapid, exponential
death at 35 C, which is averted by cycloheximide or conditions allowing no
protein synthesis. Altered phospholipid synthesis (508). Also cold sensitive
and osmotic remediable at 11°C (701). Allele T51M171 is called 25 or TS25
in references 507 and 508.
un-18: unknown-18
IR. Right of T(NM169d) and R (11%) (808).
Unknown function. Heat sensitive (507). No growth at 34°C. Growth at 25
C is substantial but not wild type, and better on complete medium than on
minimal medium. Allele T54M94 is called 41 in reference 507.
un-19: unknown-19
VR. Linked to al-3 (9%), un-9 (22%), and un-11 (14%)
(D.D. Perkins,
unpublished data).
Unknown function. Heat sensitive, 34°C versus 25°C (D.D. Perkins,
unpublished data).
un-20: unknown-20
IIR. Between ff-1 (4%) and ace-1 (15%). Right of aro-1
(5 to 9%). (816,
1052, PB).
Unknown function. Heat sensitive, 39°C versus 25°C. Best scored at
39°C on
minimal medium. Leaky. Some flat, aconidiate unpigmented growth occurs
even at restrictive temperatures (PB). Called mo(P2402t) (816, 1052).
un-21: unknown-21
IIIR. Between acr-2 and un-6 (PB).
Unknown function. Heat sensitive, 34°C versus 25°C (507). Allele
T53M26
is called 29 in reference 507.
un-22: unknown-22
VII. Linked to met-7 (1%) and un-10 (>20%) (1019,
PB).
Unknown function. Heat sensitive, growing at 20 to 28°C but not at 37°C.
Called un(61C) and un(62C). (1019).
un-23: unknown-23
VIR. Right of trp-2 (5 to 27%). Left of ws-1 (1019;
D.D. Perkins,
unpublished data).
Unknown function. Heat sensitive, growing at 20°C but not at 28°C.
Called
un(64D). (1019).
un(STL6)
See fls.
un(b39)
See un-5.
un(44409)
See un-1.
un(46006)
See un-2.
un(55701)
See un-3.
un(66204)
See un-4.
un(83106)
See un-6.
upr-1: UV photoreactivation-1
IL. Between mt (2%) and arg-1 (7%) (1094).
Sensitive to UV (273, 1096), nitrous acid (1094), ionizing radiation (940), and
nitrosoguanidine, 4-nitroquinoline 1-oxide, and ICR-170 (509). Insensitive or
marginally sensitive to methylmethane sulfonate (536). Unable to excise
dimers (1164). Normal spontaneous mutation (275). High UV-induced
mutation (273). For mutation induction by other agents, see references 509
and 940. Defective photoreactivation in vivo, but photoreactivation enzyme
functions in vitro (1094). No homozygous effect on meiosis or crossing over
(1094). Recessive in heterokaryons (979). Double mutant upr-1;uvs-3 is more
sensitive than either single mutant (1095). Double mutant upr-1;uvs-2 is no
more sensitive than the uvs-2 single mutant (506).
ure: urease
Strains carrying mutations at four distinct loci, ure-1, -2, -3, -4, lack all
detectable urease
activity (Fig. 10 and 24). Urease
from other organisms comprises numerous distinct
subunits; possibly all four Neurospora loci are structural genes (78). ure
mutation D2, which
is tightly linked to ure-1, fails to complement mutations at any of the four loci,
suggesting
a regulatory role (452). Strains carrying mutations of another type possess partial activity
but are readily scorable by poor growth on urea as the nitrogen source. These probably
represent additional loci in V (A7, S3), IV (E3, E7), or elsewhere (C5, K3, R2) (452;
H.B.
Howe, Jr., personal communication). They are not given separate entries here.
ure mutants
have been isolated by methods based on the inability to generate ammonia from urea,
using
pH indicators
(78, 452, 569). The following methods for scoring isolates are probably generally
applicable,
but all have not been tested on mutations at all loci. Method 1: Little growth on
filter-sterilized urea as the sole nitrogen source (452) (not good if amino acids must be
added). Method 2: Five- to twenty-minute scoring test, touching bits of filter paper
dipped
in urea-bromthymol blue buffer to conidia or aerial growth (570). Method 3: Color
change
when grown on slants of synthetic crossing medium (1134) containing phenol red (90
ug/ml)
and urea (3 mg/ml) as the sole nitrogen source, added before autoclaving; scored after 4
to 5 days at 34°C (PB; modified from the method of J.A. Kinsey).
ure-1: urease-1
VR. Right of ace-5 (<1%), am (1%), and ure-2 (3%).
Left of his-1 (<1%) (570, 578).
No urease activity (452, 569) (Fig. 10 and 24). Some revertants show altered heat
stability
of urease, suggesting a structural role (78). Used to study the metabolic fate of arginine
by
measuring urea accumulation (592). Used for arginine tracer experiments (217); flux
(355).
Used to determine the relative contributions of arginine and purines in urea formation
(235). For the role of urease in purine catabolism, see Fig. 24.
For scoring methods, see
ure. Possible aflele D2 fails to complement ure-1, -2, -3, or -4 (452).
Called
ure(9) (570).
ure-2: urease-2
VR. Left of am (2%). Probably right of sp. Linked to
ure-1 (3%) (570).
No urease activity (452, 569) (Fig. 10 and 24). Reversion and
complementation data
(78).
Enzyme from interallelic complementation and from some revertants shows altered heat
stability, suggesting a structural role (78). Shows hyperinducibility of purine catabolic
enzymes uricase, allantoicase, and allantoinase (872). For scoring methods, see
ure. Called
ure(47) (570).
ure-3: urease-3
IIR. Between arg-12 (7 to 12%) and ace-1 (14%) (PB). Allele B1
is closely linked to
translocation T(IR;II)B1 in the original strain. Consequently, ure-3
was at first assigned incorrectly to IR (78, 452). Point-mutant allele F29 shows linkage in
IIR, not in IR (PB).
No urease activity (452) (Fig. 10 and 24). Some revertants show altered urease
thermostability, suggesting a structural role (78). For scoring methods, see
ure.
ure-4: urease-4
IR. Left of ad-3B (3%). Probably right of his-3 (1%) (78).
(452).
No urease activity (452) (Fig. 10 and 24). Some revertants show altered heat stability of
urease, suggesting a structural role (78). For scoring methods, see ure.
ure(D2): urease
VR. At or near ure-1 ( 5) (452).
No urease activity. Fails to complement representatives of all four ure loci;
possible
regulatory gene (452).
uvs: UV sensitive
uvs and other radiation-sensitive mutants are highly pleiotropic, with
phenotypic spectra that
may (or may not) include: sensitivity to ionizing radiation, radiomimetic chemicals,
mitomycin C, or histidine; impairment of meiosis; increased frequency of deletion or
mitotic
recombination; increased
spontaneous or induced mutation; defective DNA repair; altered secretion of
deoxyribonuclease. Sensitivity is typically recessive. The most sensitive
Neurospora mutant,
uvs-2, is only 20 times more sensitive than the wild type to UV (935).
The more general name, mus (mutagen sensitive), has been adopted for
mutant loci beyond
uvs-6 (537). Several UV-sensitive mutants have names other than
uvs or mus: see gs(6), Mei-2, mei-3, nuh-4, upr-1.
Scoring is most readily accomplished by spot tests, i.e., spotting conidial suspensions on
the
surface of prepoured plates that contain sorbose (536, 932, 1023) and comparing growth
on
exposed and control plates. Properties of the uvs mutants are summarized in references
509, 537, 936, and 938. For properties of double mutants, see references 506, 539, and
1095.
uvs-1: UV sensitive-1
Not mapped.
Slightly increased sensitivity to UV (187, 273). Spontaneous and UV-induced mutation
probably normal (273, 275). Homozygous fertile and without effect on crossing over.
Ascospore viability and early growth severely impaired in homozygous crosses (187).
Reduced rate of dimer excision (1164). Difficult to score; spot tests best read early (24
h).
uvs-2: UV sensitive-2
IVR. Right of cys-4 (5%) (1023). Left of pmb (8%) (S.
Ogilvie-Villa, cited in reference
248; R. Sadler and S. Ogilvie-Villa, personal communication) and the
T(S4342) right
breakpoint. Linked to T(AR209), T(T54M50), and T(ALS179) (2
to 6%), which mark the
IVR tip (808).
Sensitive to UV (273, 1023), ionizing radiation (537, 935, 940), methyl methane sulfonate
(536, 537), nitrosoguanidine (509, 935), mitomycin C (537), 4-nitroquinoline 1-oxide
(509),
nitrous acid (D.R. Stadler and E. Crane, personal communication), and ICR-170 (509).
Slight or no sensitivity to histidine (537, 759). No dimer excision (1164). Normal
spontaneous mutation (275). High UV-induced mutation rate (273); for mutation
induction
by other agents, see references 509 and 940. Homozygous fertile; no effect on meiosis or
crossing over. Recessive in heterokaryons (1023). uvs-2 is the most UV
sensitive of Neurospora mutants (15 to 20 times wild type) (537, 938). Used
to show that
DNA repair is induced by a small dose of UV (1022). Used to demonstrate
postreplication
repair (130). Only known allele was discovered in several Seattle stocks of mixed
ancestry,
and thus may be present in lab
stocks elsewhere (1023; D.R. Stadler, personal communication). Not to be confused with
a cytoplasmically determined UV-sensitive mutation called uvs-2 in reference
187, but now
called [uvs(cyt)].
uvs-3: UV sensitive-3
VL. Linked to cys-10 (3 to 7%), probably to the left (538, 932).
Allele ALS11 is sensitive to UV (273, 932, 933), ionizing radiation (537, 933, 940),
methyl
methane sulfonate (536), nitrosoguanidine (509, 933), mitomycin C (195, 537), histidine
(932), and 4-nitroquinoline 1-oxide and ICR-170 (509). Reverts spontaneously (932). No
UV-induced mutation (273). For mutation induction by other agents, see references 509
and 940. Increased spontaneous mutation (275, 537). Dimer excision delayed and at
reduced rate (1164). Defective photoreactivation in vivo, but photoreactivation enzyme
functions in vitro (934). Defective in extracellular nuclease, giving reduced halo around
colonies on DNA agar (538). Apparently deficient in proteolytic conversion
of nuclease precursor to active intra- and extracellular deoxyribonucleases, but this effect
could be indirect (360). Increased stability of CPS(Pyr) and ACT activities in vitro also
suggests that protease activity may be reduced in uvs-3 mutants (882). Causes
increased
duplication instability (mitotic recombination or deletion or both) (932). Conidial
viability
is low (275, 932). Double mutant upr-1;uvs-3 is much more sensitive to UV
than is either
single mutant (1095). Double mutant uvs-3;uvs-6 is inviable (506).
Homozygous barren,
with block before karyogamy (860, 932). Shows high level of repair of genetic damage
without induction in rescuing a heterokaryotic component that carries
potentially lethal mutagen-induced damage (1022). Probable allele FKO16, isolated as
halo
mutation nuh-4, resembles uvs-3 allele ALS11 but is less extreme in
some properties, e.g.,
it shows better conidial survival (538; see reference 537).
uvs-4: UV sensitive-4
IIIR. Left of ad-4 (4%) (932).
Sensitive to UV (273, 932, 933) and histidine (932). Moderately sensitive to methyl
methane
sulfonate (537). Insensitive or only slightly sensitive to nitrosoguanidine (537, 933); no
increased sensitivity to ionizing radiation (940) or mitomycin C (195, 537). Probably
normal
spontaneous mutation (275). Reduced UV-induced mutation (273). For mutation
induction
by other agents, see reference 940. Homozygous fertile; no effect on intragenic or
intergenic
recombination. Ascospore viability and early growth severely impaired in homozygous
crosses (932). Recessive in heterokaryons (979).
uvs-5: UV sensitive-5
IIIR. Linked to vel (1%) (932).
Sensitive to UV (273, 932, 933), nitrosoguanidine and ICR-170 (509), and histidine (932);
not sensitive to ionizing radiation (940) or 4-
nitroquinoline 1-oxide (509). Slow growth. Normal dimer excision (1164). Spontaneous
mutation normal (275); UV-induced mutation reduced (273). For mutation induction by
other agents, see references 509 and 940. Homozygous barren (932) with meiosis
blocked
at pachytene (860).
uvs-6: UV sensitive-6
IR. Between thi-1 (3 to 8%) and ad-9 (4%) (538; E. Kafer,
personal communication; D.
Newmeyer, unpublished data). Very close to met-6 (< 1%) (D. Newmeyer,
unpublished
data). (937).
Increases sensitivity to UV (273, 937), ionizing radiation (537, 937, 940), nitrosoguanidine
(509, 537), histidine (755), methyl methane sulfonate (536), ICR-170 and 4-nitroquinoline
1-oxide (509), and possibly mitomycin C (537). Normal UV-induced mutation (273).
For
mutation induction by other agents, see references 509 and 940. Increased spontaneous
mutation not evident in the ad-3 system (275), but is found for recessive
lethals in a
heterokaryon test system (E. Kafer, personal communication). Normal dimer excision
(1164). Defective in extracellular nuclease, giving reduced halos around colonies on
DNA
agar (538). Increased duplication instability due to mitotic recombination, deletion, or
both
(759). Homozygous barren (759), with a block at crozier differentiation (860). Reduced
conidial viability (537). Switches to stop-start growth after initial normal growth (D.
Newmeyer, unpublished data). Not completely recessive in heterokaryotic
conidia (979). Increased stability of CPS (Pyr) activity in vitro suggests that protease
activity
may be reduced in the mutant uvs-6 (882); see also reference 360. Double
mutant
uvs-3;uvs-6 is inviable (506).
uvs(FKIO4)
See mus-9.
val: valine
VR. Right of at (10%). Linked to ilv-2 (0/135) (PB).
Strains carrying alleles 33026 and 33050 appear to be valine auxotrophs, not requiring or
responding to added isoleucine (482, PB). Not defective in valyl-tRNA synthetase (J.
Evans,
personal communication via J.A. Kinsey), as are the only known val mutants
of bacteria.
Requirement somewhat leaky; not temperature sensitive. Relation to ilv-1 and
ilv-2 is
uncertain. An ilv mutant strain with an incomplete isolation number,
ilv(?6201), was
incorrectly designated val (45201) and was the source of linkage data for a
locus
erroneously shown as val on maps made before 1980.
van: vanadate resistant
VIIL. Left of nic-3 (4%) (B.J. Bowman, personal communication).
Resistant to vanadate (0.1 to 1.0 mM) in Vogel minimal medium with 0.15 mM
phosphate
and 20 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) buffer,
adjusted
to pH
7.5 (B.J. Bowman, personal communication).
var-1: variant-1
Unmapped. Single-gene difference.
Slow growth (62% normal). Aerial hyphae decreased to give shaven appearance.
Functional protoperithecia absent. Lysed areas appear
and spread in old cultures. Not rejuvenated in heterokaryons. Originated in experiment
on
uninterrupted growth. (86).
vel: velvet
IIIR. Linked to phe-2 (1%). Right of T(D305) and ro-2
(6%). Left of tyr-1 (3 to 5%) (814, 816, PB).
Soft, colonial growth habit (789). Flat at first; then becomes elevated. Eventually
conidia
may be formed in aerial puffs. Good female
fertility. col-13 (R2471) and col-15 (R2531) are putative alleles.
vis(3717): visible
Linked to the right of the centromere in linkage group I (482, 486), but inseparable from
translocation T(I;III)3717 (808). No corresponding
point mutation is known.
Aconidial flat morphology.
wa: washed
VR. Linked to inl (6%) (812).
Thin, spreading surface growth and conidiation (812). Called spco-2.
wc-1: white collar-1
VIIR. Right of met-9 (1 to 4%). Left of un-10 (7%) and
for (6%) (724, 812, 816).
Carotenoids absent from mycelia; conidia become pigmented with some delay. Named
because nonconidiating rim at top of agar slant remains white. A double mutant with
fl or
other nonconidiating mutant would be classed as albino. Regulatory mutants for
photoinduced carotenogenesis via blue light receptor might be expected to have a similar
phenotype (444, 445). A blue light treatment (given in vivo), which increases the activity
of soluble and microsomal enzymes required for phytoene biosynthesis in the wild type,
does
not do so in the mutant wc-1 (445). Fails to show phototropism of perithecial
beaks
when
used as the female (protoperithecial) parent, but not when used as the male (fertilizing)
parent (R. W. Harding, personal communication). Useful genetic marker (725, 800).
Scoring clearest at high temperatures
(34 C).
wc-2: white collar-2
IR. Right of T(NM103), T(ALS182), and thi-1 (8%). Left of
ad-9 (7 to 20%) (PB).
Resembles wc-1. Detected in a glp-1 strain (J.B. Courtright,
personal communication); wc-2
is separable from glp-1 by crossing over (PB).
ws-1: white spore-1
VIR. Right of trp-2 (38%) (822).
Delayed ascospore maturation; autonomous. Ascospores fail to darken or do so slowly.
Black spots appear on some ws-1 ascospores. In aged crosses,
a few percent of the ws-1 ascospores darken and are capable of germination.
Fertile and
prototrophic, with normal tyrosinase activity vegetatively. Photograph of asci. (822).
Second-division segregation frequencies may be as high as 80 to 96% (586, 822),
providing
evidence for chiasma interference.
ws-2: white spore-2
VI. Linked to ylo-1 (16%), trp-2 (2%) (A. Kruszewska, personal
communication), and the
centromere (9 to 24% in ordered asci) (586). Recombines with ws-1 (R.L.
Phillips, personal
communication). (816).
Ascospores white initially, browning with age. Autonomous (816).
xdh-1: xanthine dehydrogenase-1
II. Linked to pe (14%) and alc-1 (24%) (872).
Defective in purine catabolism. Unable to use hypoxanthine as the sole nitrogen source.
Lacks xanthine dehydrogenase (xanthine oxidase) (872) (Fig.
24). nit-1, -7, -8,
and -9 mutants
are also deficient in xanthine dehydrogenase activity because of a defect in the
molybdenum
containing cofactor that is common to xanthine dehydrogenase and nitrate reductase
(741,
1080, 1081).
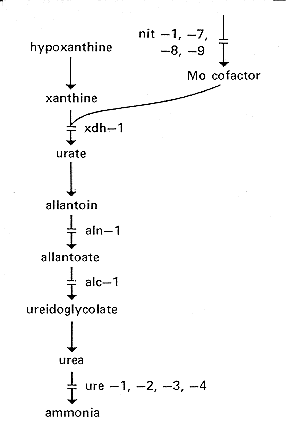
FIG. 24. Purine catabolic pathway, showing the sites of action of the
xdh, aln, alc, and ure
genes (452, 569, 872). The molybdenum cofactor is essential to both xanthine
dehydrogenase and nitrate reductase
ylo: yellow
Yellow carotenoids. In addition to strains with mutations at loci designated
ylo, strains with
some al-1 alleles (especially ALS4 and RES-25) produce lemon-yellow
carotenoids, although
in reduced quantity (1039, 1042, 1071).
age-3 mutants are also yellow. Many morphological mutants appear to be
abnormal in
carotenoid metabolism and may appear yellowish or pale rather than the wild-type
orange.
ylo-1: yellow-1
VIL. Between cys-1 (8%) and ad-1 (6%). Probably right of
Bml (2%) (1012, PB). (381).
Yellow carotenoids (381). Affects synthesis of neurosporaxanthin
(4'-apo-beta'-caroten-4'-oic acid); citations in reference 398. Lesion probably involves
the
conversion of lycopene to 3,4-dehydrolycopene or the conversion of either torulene or
gamma-carotene to neurosporaxanthin (398 and references therein) (Fig. 9). Resembles
the
orange wild type in young cultures, but color differences become clear with age.
Expressed
in both conidia and mycelia. Undefined modifiers affect intensity. Fails to complement
with many of the al-1 and al-2 albino strains (R.E.
Subden, personal communication).
ylo-2: yellow-2
IL. Right of In(H4250); hence, of suc. Left of arg-1 (1%)
(816).
Rather sparse yellow conidia (816). No information on carotenoids; possibly the
yellowish
color is a secondary effect of abnormal development. Inferior to ylo-1 as a
marker. First
shown distinct from ylo-1 by A.M. Kapular (via FGSC).
ylo-3: yellow-3
IIR. Probably allelic with fl (0/56) (D.D. Perkins, unpublished data). Listed
as IIIR; no
data given (1040). Not linked to ylo-1 or ylo-2 (A.M. Kapular, via
FGSC).
Pale yellow. Slow to conidiate. (A.M. Kapular [via FGSC], D.D. Perkins, unpublished
data).
Return to the FGSC Home page

Contact the FGSC
Last modified 4/24/96 KMC


