
Fungal Genetics Stock Center.
The simplest way to obtain cultures of Neurospora is to contact the FGSC at http://www.fgsc.net/.
Thousands of mutant stocks can be viewed on the web site. The meanings of the
mutant classes can be looked up in the Neurospora
Compendium: Perkins, D. D., A. Radford, and M. S. Sachs. 2001. The
Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego. (323 pp.) A
useful earlier version is available on line at http://www.fgsc.net/compend/compend.html
Order by email. Strains are mailed out to customers, arriving usually between one to two weeks of ordering. Invoices are included with the strains. (The FGSC asks a small fee per strain, but this fee can be waived in the case of financial hardship.)
Other collections and repositories
The American Type Culture Collection
Centraalbureau voor Schimmelcultures
Deutsche Sammlung von Mikroorganismen und
Zellkulturen GmbH
Local universities and colleges
Many universities and colleges use Neurospora in labs and might be willing to give
subcultures of their stocks. If a local biology or genetics department has a
Neurospora researcher they will probably be happy to give you some strains to
get you started.
Collect wild types from nature
In warmer areas of the world Neurospora can be obtained by plating soil samples
using a special medium (see detailed protocol section). In temperate regions
Neurospora can sometimes be found after burns eg under tree bark after a forest
fire, or at camp fire sites.
This section offers a brief overview and more detailed set of protocols for growing and handling Neurospora.
For details of Neurospora protocols consulted one of the following:
-- Full Neurospora
protocols
-- Streamlined set of
basic protocols assembled by D E A Catcheside at Flinders University, Adelaide,
Australia
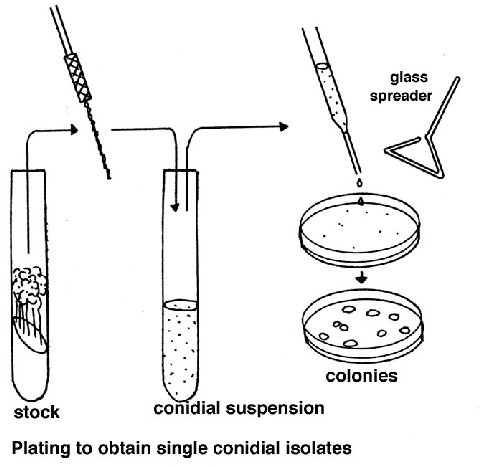
Culturing Cultures for routine use are usually grown in tubes or
plates containing a medium consisting of Vogel’s salt solution
Sucrose
Agar
Water
Details of this recipe here.
A new medium was described by R. Metzenberg in FGN 51

Plating
Cell populations (conidia or ascospores) are plated on a medium consisting
of Westergaard and Mitchell salt solution Sorbose + fructose + glucose OR
sorbose + sucrose mixture (sorbose restricts growth)
Agar
water
Check the full protocol section for quantities.
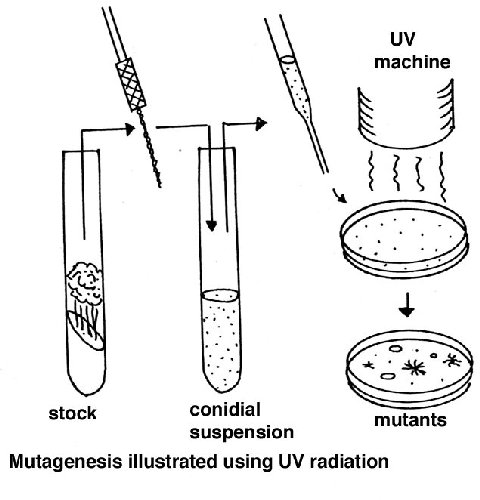
Random somatic mutations
A suspension of macroconidia is exposed to a mutagen such as nitrous acid or
ultraviolet light, using a dose that gives about 50% survival (this dose needs
to be found first, using the equipment/chemicals available). Since conidia have
several nuclei, this dose results in a large proportion of cells with one
nucleus, allowing expression of recessive mutations. Alternatively microconia
can be used, but preparing a sample of these is more laborious. After plating,
the screens or selections can be applied for the specific mutant colony
phenotype required.
Mutating one specific gene
RIP
A convenient molecular method is to use RIP (repeat-induced point mutation). A
clone of the wild type copy of the gene to be mutated is transformed into a
wild type. The introduced strain will probably insert ectopically. The
resulting strain is crossed to the opposite mating type. Both copies of the
original transformed strain will be RIPPed, and will contain multiple GC - >
AT mutations. Most RIPPed alleles represent functional knockouts.
Replacement.
An engineered mutant allele is inserted at the resident locus of that gene by
homologous recombination. The new allele can be a complete functional knockout
or have partial function.
Specific locus test.
To obtain new mutations of your favorite gene (yfg), mutagenize the following
heterokaryon
( aux-1 yfg aux-2+ + aux-1+ yfg+ aux-2)
Screen conidia for yfg phenotype.
Crossing
Here again a brief overview is presented, whereas a fuller account
can be found in the Adelaide document.
Parents of opposite mating type are combined in tubes or on plates containing a
special crossing medium consisting of
Westergaard and Mitchell nutrient solution
Sucrose
Agar
Water
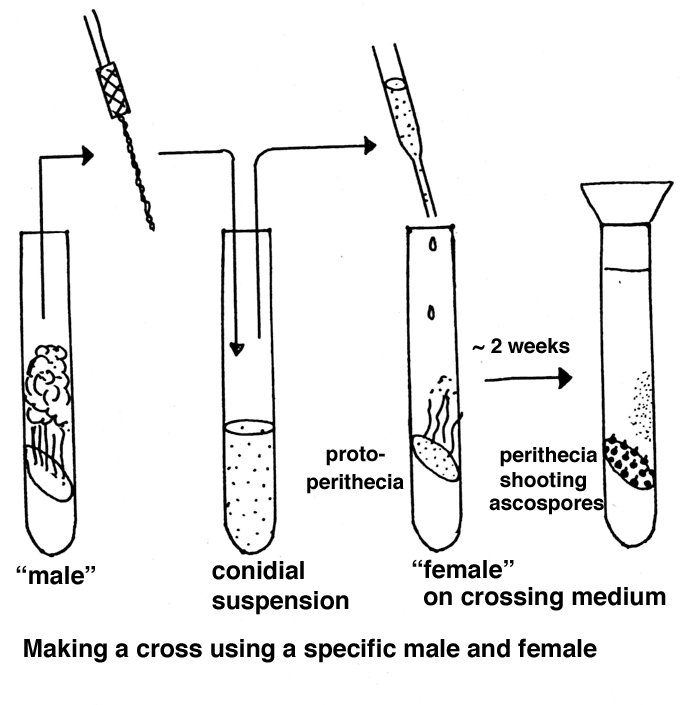
If one strain is to be used as maternal parent, that strain is grown first and after a week the paternal strain is added as a conidial suspension.
Rough chromosomal location of a new mutant can be obtained by crossing
it to special mapping strains such as multicent or alcoy. Map position can be
narrowed down by crossing to other markers in the general vicinity. Special
RFLP mapping strains are useful in this regard. Check the maps and stock lists
at FGSC.
Complementation test
To see if two mutants (m1 and m2) complement, they are combined in a
forced heterokaryon on minimal medium (aux-1 and aux–2 are auxotrophic forcing
markers):
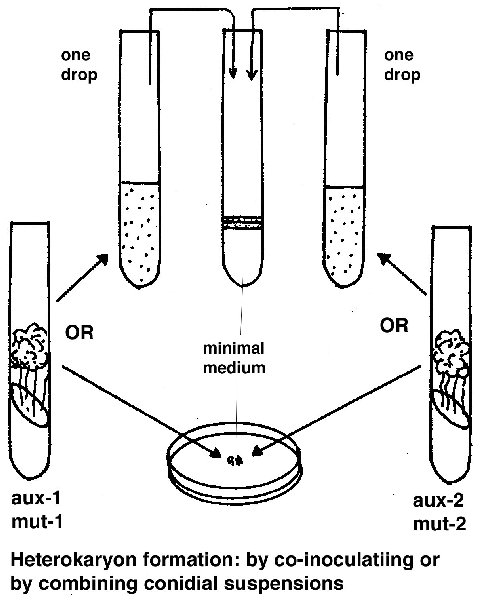
(aux-1 m1 aux-2+ + aux-1+ m2 aux-2)
if the phenotype is wild type, then the mutants complement and are not allelic.
Cloning by functional complementation
(= mutant rescue).
The clones of a library are tested to see if they can restore wild type
function to a mutant culture. The total set of clones is gradually narrowed
down to find the clone of the gene of interest.
Positional cloning.
If the rough position of a gene is known, then it is possible to zero in on
candidate genes known from the genomic sequence to be in that vicinity. Cosmid
clones used in assembling the genome sequence can be tested individually and
eventually subdivided to find the right gene.
Polymerase chain reaction
Primers designed from the genomic sequence are used to chemically amplify the
desired gene in a test tube.
Genetic engineering
Transgenic strains are made by transforming recipient cells with a
vector containing the desired transgene(s) plus a selectable marker.
Integration is almost always ectopic. (No autonomously replicating plasmids are
available at present.)
Staining cells, chromosomes, organelles, etc.
A more detailed look at cytology and genetics in
Neurospora
N.B. Raju
A brief guide to cytogenetic staining methods for Neurospora
N B Raju, Stanford University.
Fungal nuclei and chromosomes are relatively small but they are readily visualized by light microscopy after appropriate staining. Iron-hematoxylin is excellent for routine cytology of Neurospora and other fungi. It stains the chromosomes, nucleolus, spindle, spindle-pole bodies, and ascus apical pore very clearly (Lu and Raju 1970; Raju and Newmeyer 1977; Raju 1980). However, it is less useful for pachytene chromosome analysis because the chromomere detail is not clearly revealed or because the large dark-staining nucleolus may obscure underlying chromosomes. Aceto-orcein is very good for pachytene chromosome analysis but does not stain spindles, spindle-pole-bodies or the nucleolus satisfactorily (see Singleton 1953; Barry 1966, 1996). While these two staining methods work well in the hands of experienced cytologists, they are somewhat unpredictable because of many variables involved in the preparation of stains and their use.
Fluorescent staining of Neurospora nuclei is quick, easy, and much simpler than conventional methods. DAPI and Hoechst 33258 are the simplest and quickest to use for nuclear counts in the conidia and hyphae (Raju 1982). Acriflavin is very good for detailed observations on chromosomes during ascus development (Raju 1986). For observations, an epifluorescence microscope fitted with a mercury lamp is very versatile because of its wide-range emission spectrum including near-UV. The near-UV excitation is essential for DAPI and Hoechst 33258. Acriflavin uses blue-light excitation. In addition to the light source, one or more sets of specifically matched excitation filter / dichroic mirror / barrier filter combinations are needed to deliver the radiation of desired wavelength to the specimen and to transmit the resulting fluorescence to the observer.
In the last few years, GFP-tagged histone H1 and b-tubulin genes have greatly aided in the visualization of nuclei and microtubules in the conidia, hyphae, asci, and ascospores. In fact, any gene of interest can now be tagged with GFP, and its localization and temporal limits of expression may be visualized with a fluorescence microscope (Freitag et al. 2004). Any research microscope (e.g. Zeiss, Leitz, Nikon, or Olympus) equipped with epifluorescence illumination can be tailored for GFP observations with an appropriate filter set specifically suited for a variant of GFP (sGFP). Our Nikon Microphot FX microscope is fitted with a filter set from Chroma #41012 (480/40 nm excitation filter, 505 nm dichroic mirror, 510 nm long pass emission filter).
FM1-43 and DASPMI (stain mitochondria), and FM4-64 (stains plasma membrane and organelle membranes) have been used as counter stains for observations of mitochondria and GFP-tagged nuclei in live-cell imaging of growing hyphae using confocal microscopy (Hickey et al. 2002).
References
Barry, E. G. 1966. Cytological techniques for meiotic chromosomes in
Neurospora. Neurospora Newsl. 10: 12-13.
Barry, E. G. 1996. Fungal chromosomes. J. Genet. 75: 255-263.
Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker, and N. D. Read. 2004. GFP
as a tool to analyze the organization, dynamics and function of nuclei and
microtubules in Neurospora crassa (In preparation).
Hickey, P. C., D. J. Jacobson, N. D. Read, and N. L. Glass. 2002. Live-cell
imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet.
Biol. 37: 109-119.
Lu, B. C., and N. B. Raju. 1970. Meiosis in Coprinus. II. Chromosome pairing
and the lampbrush diplotene stage of meiotic prophase. Chromosoma 29: 305-316.
Perkins, D. D., N. B. Raju, E.G. Barry, and D. K. Butler. 1995. Chromosome
rearrangements that involve the nucleolus organizer region in Neurospora.
Genetics 141: 909-923.
Raju, N. B. 1978. Meiotic nuclear behavior and ascospore formation in five
homothallic species of Neurospora. Can. J. Bot. 56: 754-763.
Raju N. B. 1980. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol.
23: 208-223.
Raju, N. B. 1982. Easy methods for fluorescent staining of Neurospora nuclei.
Neurospora Newsl. 29: 24-26.
Raju, N. B. 1986. A simple fluorescent staining method for meiotic chromosomes
of Neurospora. Mycologia. 78: 901- 906.
Raju, N. B., and D. Newmeyer. 1977. Giant ascospores and abnormal croziers in a
mutant of Neurospora crassa. Exptl. Mycol. 1: 152-767.
Singleton, J. R. 1953. Chromosome morphology and the chromosome cycle in the
ascus of Neurospora crassa. Am. J. Bot. 40: 124-144.